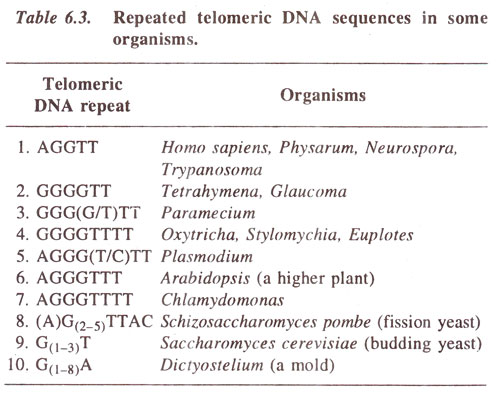
Fig. 6.12. Ultrastructure of chromosome (drawn from an electron micrograph. Du Praw : Cell and Molecular Biology).
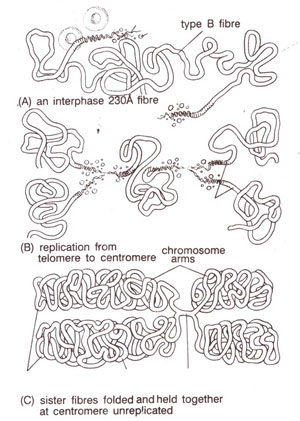
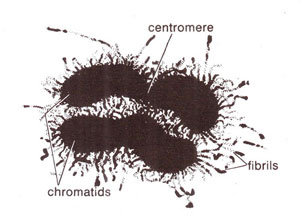
Fig. 6.13. Structural organization in a chromosome according to folded fibre model (redrawn from Du Praw : Cell and Molecular Biology).
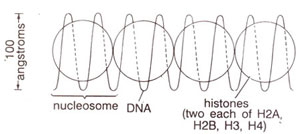
Fig. 6.14. Original proposal for nucleosome repeating units, with DNA wound on a series of beads. Actual shape of histone complex or the path of DNA was not known, (redrawn from Sci. Amer. Vol. 244, 1981).
In the past, study of chromosome ultrastructure was an important area, where electron microscope had failed to give us a clear picture of organization of DNA. For the study of chromosomes with the help of electron microscope, whole chromosome mounts as well as sections of chromosomes were studied. Such studies had demonstrated that chromosomes have very fine fibrils having a thickness of 2 nm-4 nm (Fig. 6.12). Since DNA is 2 nm wide (consult
Chemistry of the Gene 1. Nucleic Acids and Their Structure), there is possibility that a single fibril corresponds to a single DNA molecule. However, whether these fine fibrils represent the natural state of DNA or is an artifact was not known. Moreover, the structure corresponding to protein component of the chromosomes was also not known, although the position of proteins was differently shown in different chromosome models.

Fig. 6.12. Ultrastructure of chromosome (drawn from an electron micrograph. Du Praw : Cell and Molecular Biology).
A meaningful picture of the structure of chromosome could be earlier drawn from available data on ultrastructure of eukaryotic chromosomes. If a single chromatid has a single long DNA molecule, as has been proved, we have no choice but to believe that DNA should be present in a coiled or folded manner. This can also be inferred from the fact that the thickness of a chromosome is usually hundred times that of DNA and the length of DNA found in a chromosome is several thousand times the length of the chromosome.
The manner of coiling and folding of DNA was a matter of debate and dozens of important models for this purpose were available. It is not desired here to present these different models, or else to discuss their merits or demerits. A popular model was the
folded fibre model (Fig. 6.13) given by
E.J. Du Praw in 1965. The most important and universally accepted model is
nucleosome model proposed in 1974. This model involving nucleosome concept will be described briefly here; its detailed account will be given later in
Organization of Genetic Material 1. Packaging of DNA as Nucleosomes in Eukaryotes.

Fig. 6.13. Structural organization in a chromosome according to folded fibre model (redrawn from Du Praw : Cell and Molecular Biology).
Structure of chromosomes at molecular level (a) Nucleosome-subunit of chromatin. In 1974,
R.D. Kornberg and
J.O. Thomas proposed an attractive model for basic chromatin structure involving DNA and histones. They suggested that DNA interacts with a tetramer (H3
2-H4
2) and two molecules of an oligomer (H2A-H2B), so that a tetramer involving two molecules each of the histones H3 and H4, is associated with two molecules each of the histones H2A and H2B and with 200 base pairs of DNA. This makes a repeating unit (Fig. 6.14). One molecule of H1 is also associated with each repeating unit. They also proposed that the tetramer makes the core of the unit and oligomers determine the spacing thus giving a flexible structure.
This model is supported by biochemical and electron microscopy results.
P. Oudet et at (1975) proposed the term
nucleosome for repeating units which were observed as beads on strings (or string on beads—see
Organization of Genetic Material 1. Packaging of DNA as Nucleosomes in Eukaryotes) under electron microscope.

Fig. 6.14. Original proposal for nucleosome repeating units, with DNA wound on a series of beads. Actual shape of histone complex or the path of DNA was not known, (redrawn from Sci. Amer. Vol. 244, 1981).
Nucleosome (12.5 nm in diameter) = 200 base pairs + 2 molecules each of H2A, H2B, H3, H4
Chromatin structure and the nucleosome have been dealt with, in much greater detail in
Organization of Genetic Material 1. Packaging of DNA as Nucleosomes in Eukaryotes, after discussing the structure and synthesis of nucleic acid in
Chemistry of the Gene 1. Nucleic Acids and Their Structure and
Chemistry of the Gene 2. Synthesis, Modification and Repair of DNA.
(b) Telomere. In recent years the structure of telomeres in a wide variety of organisms has been studied to demonstrate that telomeres are highly conserved elements throughout the eukaryotes, both in structure and function. Telomeric DNA has been shown to consist of simple randomly repeated sequences, characterized by clusters of G residues in one strand and C residues in the other. Another feature is a 3' overhang (12-16 nucleotides in length) of the G-rich strand. Some of the telomeric DNA sequences found in eukaryotes are given in Table. 6.3.

Fig. 6.15. A metaphase plate of mitotic chromosomes, hybridized in situ with a probe having telomeric DNA repeat sequence. The telomeric sequence can be seen at the ends of all chromosomes.
The same repeated sequence is found at the ends of all chromosomes in a species (Fig. 6.15) and the same telomere sequence may occur in widely divergent species, such as humans, and some acellular slime molds
(trypanosomes) and fungi like
Neurospora. At every telomere, as much as 10 kilobases of this repeat sequence may occur. The telomeric DNA is also complexed with non-histone proteins, the complex structure being associated with nuclear lamina, as shown in
Oxytricha, a ciliated protozoan. The telomeric DNA is synthesized under the influence of telomerase, an enzyme which has been shown to be a ribonucleoprotein, whose RNA component works as a template for synthesis of telomeric DNA repeats (for more details consult
Chemistry of the Gene 2. Synthesis, Modification and Repair of DNA).

Fig. 6.15. A metaphase plate of mitotic chromosomes, hybridized in situ with a probe having telomeric DNA repeat sequence. The telomeric sequence can be seen at the ends of all chromosomes.