Fig. 15.2. The gal operon of E. coli, showing three structural genes and the promoter.
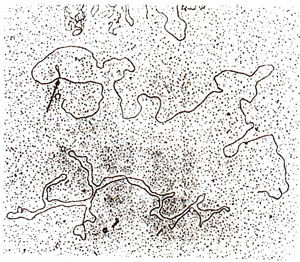
Fig. 15.3. Diagrammatic representation of an electron micrograph of λdgal+/λdgalm DNA heteroduplex, showing a loop representing an IS in λdgalm
In prokaryotic as well as eukaryotic genomes, certain sequences are capable of moving from one site to another and are described as
transposable elements (TE). These are sometimes considered as synonymous to the entities to be described later in this section as transposons. According to some workers, these TE may include (i) insertion sequences or
IS elements, (ii) transposons, also characterized by certain structural features other than their ability to transpose and (iii)
retroelements which involve reverse transcription, either in their origin or propagation, insertion sequences or IS elements were the first transposable elements, identified as spontaneous insertion in some bacterial operons.
These insertions inactivate the gene and do not allow transcription and translation of this and the following (other genes downstream) genes in the operon. The first operon, where IS elements were detected, was
gal operon of
E. coli (Fig. 15.2), responsible for synthesis of three enzymes (epimerase, transferase, kinase) needed for metabolism of gajactose (for operon model, consult
Regulation of Gene Expression 1. Operon Circuits in Bacteria and other Prokaryotes).

Fig. 15.2. The gal operon of E. coli, showing three structural genes and the promoter.
These mutations caused by IS elements were called polar mutations, since transcription from all sequences downstream from insertion site was inhibited. Since these mutations did not revert back, and because different mutagens did not affect frequency of reversion, these were neither deletions, nor frameshift or point mutations (for mutations including frameshift, consult
Mutations : 1. Morphological Level (Including Lethal Mutations),
Mutations : 2. Biochemical Level (Biochemical and Microbial Genetics) and
Mutations: 3. Molecular Level (Mechanism)). In order to study the mechanism of induction of polar mutations, lambda (λ)phage (which picks up
gal region after inserting in this region) was used. λ
dgalm was isolated from mutant
galm and its -DNA was used for synthesis of radioactive RNA. Certain fragments of this RNA hybridized with
galm, but not with
gal+, suggesting that an extra piece of DNA was present in
galm.
These specific fragments also hybridized with DNA from other polar mutants, suggesting that the same piece of extra DNA is found inserted in all polar mutants. DNA from λ
dgalm was hybridized with that from λ
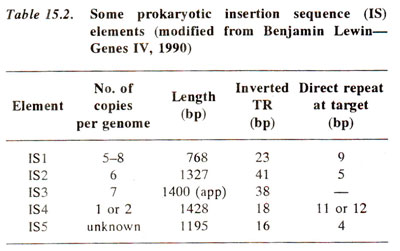
gal+, and extra piece of DNA (768 bp) could be located in the from of a loop under an electron microscope (Fig. 15.3) and was designated as IS1. Several such IS elements were later detected in
E. coli (Table 15.2). All of them are characterized by (i) the presence of inverted terminal repeats (TR) required for transposition (see later in this section); (ii) the ability to generate at the target site direct repeats of flanking DNA; and (iii) presence of open reading frames, coding for enzyme transposase, which is essential for transposition.

Fig. 15.3. Diagrammatic representation of an electron micrograph of λdgal+/λdgalm DNA heteroduplex, showing a loop representing an IS in λdgalm