Mechanism of Transposition

Fig. 15.21. Structure of a transposon showing presence of transposase gene and a regulator gene.
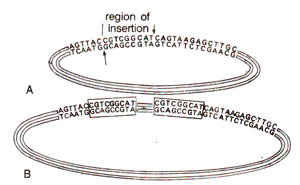
Fig. 15.22. The origin of inverted repeats due to insertion of a segment after cleavage of DNA strands at two different places separated by nine nucleotides. 'A' shows the original plasmid undergoing staggered cleavage and B is the resultant after insertion.
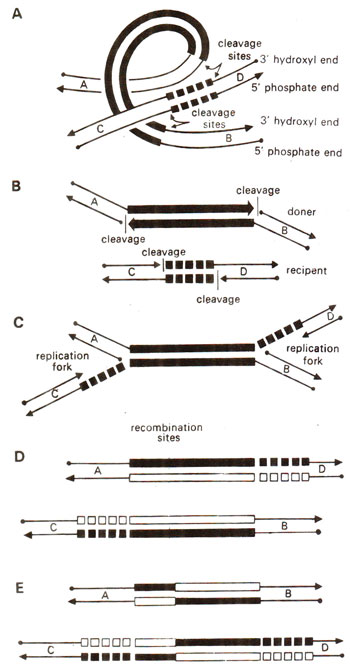
Fig. 15.23. A suggested pathway involving five steps (A, B, C, D, E) to explain transposition. The donor DNA includes a transposon (shown by thick bars), and cleavage takes place at the ends of transposon sequence in two strands of donor DNA and also at the two ends of target nucleotide sequence in two strands of recipient DNA. Both the transposon and target sequences are duplicated which is followed by reciprocal recombination (redrawn from Sci. Amer. Vol. 242, 1980).
It is also speculated now that the multiple drug resistance of plasmids has evolved due to collection of transposons, each carrying a gene for resistance against one or more antibiotics (Fig. 15.22). Since transposon like Tn3 can move among quite unrelated plasmids or bacteria, identical transposons can be found in different bacterial species.
In addition to their ability to join unrelated DNA segments, the transposable elements can also promote rearrangement and deletion of genetic material. In this category, we include a phage called Mu or mutator, which has a property of inserting at multiple sites in host bacterium thus causing different kinds of mutations. But Mu can also exist an an infectious virus. In the virus particle, Mu DNA is sandwiched between two short segments of bacterial DNA, which Mu picked up from bacterial chromosome. When Mu infects new cell, it sheds old bacterial DNA and is transposed to a site in the new host chromosome. It has also been shown that Mu can catalyse a remarkable series of chromosome rearrangements like: (i) fusion of two DNA molecules e.g. bacterial DNA and phage DNA, (ii) deletion of an adjoining DNA segment, (iii) inversion in adjoining region, and (iv) transposition of a segment of bacterial DNA to plasmid.
Through a detailed study of nucleotide sequence of insertion sequence element IS1, it could also be shown that insertion of the element is accompanied with duplication of a sequence 5, 7, 9 or 11 nucleotides long, on either side of insertion sequences. The origin of these repeats is explained to be due to staggered cleavage,

Fig. 15.22. The origin of inverted repeats due to insertion of a segment after cleavage of DNA strands at two different places separated by nine nucleotides. 'A' shows the original plasmid undergoing staggered cleavage and B is the resultant after insertion.

Fig. 15.23. A suggested pathway involving five steps (A, B, C, D, E) to explain transposition. The donor DNA includes a transposon (shown by thick bars), and cleavage takes place at the ends of transposon sequence in two strands of donor DNA and also at the two ends of target nucleotide sequence in two strands of recipient DNA. Both the transposon and target sequences are duplicated which is followed by reciprocal recombination (redrawn from Sci. Amer. Vol. 242, 1980).
- what is the mechanism of recognition of the inverted repeat ends of transposable genetic elements?
- what proteins other than those coded by the transposon are needed for transposition?
- what are the additional genetic aspects of the regulation of transposition?
The insertion of a transposable genetic element in one orientation will switch off a specific gene and switch on another gene. A reverse effect may be - observed due to insertion in a different orientation. It is in this respect that transposons and other transposable genetic elements in bacteria and higher organisms control gene regulation.




