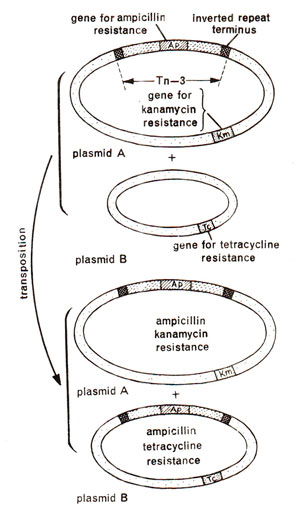
Fig. 15.5. Transposition of a transposon (Tn3) carrying ampicillin resistance (Ap)from one plasmid (plasmid A carrying additional resistance for kanamycin, Km)to another plasmid (plasmid B carrying tetracycline resistance Tc).
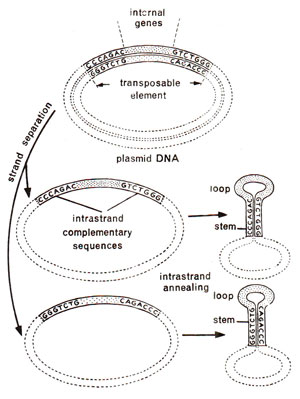
Fig. 15.6. Presence of inverted repeats at the two ends of each strand of a transposon in a plasmid and the formation of characteristic stem and loop in each strand on strand separation due to denaturation. Stem and loop formation is an evidence for inverted repeats.
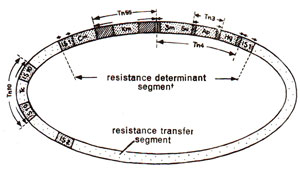
Fig. 15.7. A plasmid with several transposons of varying sizes. Note the presence of a smaller transposon within a bigger transposon.
Transposon is a term used in 1974 by
R.W. Hedges and
A.E. Jacob of Hammersmith Hospital in London, for a DNA segment or genetic element which could move from one molecule to another and carried resistance for antibiotic ampicillin. They observed that transfer of such antibiotic resistance from one plasmid to another is accompanied by an increase in size of the recipient DNA molecule (or plasmid). This recipient plasmid could donate this resistance to another plasmid, which also showed a similar increase in size thus proving that transfer of a DNA segment was involved. This DNA segment, called a
transposon, can occupy different sites in the genome, can be transposed between two bacterial chromosomes or two plasmids or between a plasmid and a bacterial chromosome. It was also shown that transfer of
transposon carrying ampicillin resistance could take place not only in plasmids as shown in Figure 15.5, but even in those bacteria which were mutant for the gene
rec A responsible for recombination, thus suggesting that these transfers could not involve normal recombination process. Properties of some of the known transposons are given in Table 15.3.

Fig. 15.5. Transposition of a transposon (Tn3) carrying ampicillin resistance (Ap)from one plasmid (plasmid A carrying additional resistance for kanamycin, Km)to another plasmid (plasmid B carrying tetracycline resistance Tc).
It was also shown that two ends of each of the two
strands in transposon consisted of hucleotide sequences that were complementary to each other, but in reverse order. For instance if one end had CCCAGAC, the other end will have GTCTGGG. These inverted repeats will form stem and loop structure characteristic of such repeats. When a plasmid
with transposon was denatured (heated to separate two strands of DNA) and each single strand was allowed to base pair among its ownself,
such stem and loop structures were actually observed (Fig. 15.6). All transposons studied so far have been found to consist of inverted repeats at the two ends. These repeated sequences may range from few nucleotides to as many as 1400.
It has been shown that insertion of any gene between two transposons makes the transfer of this gene possible without normal recombination process. Therefore, it has been seen that genes other than those for antibiotic resistance which are known to be absent in transposons have been found to accompany transposition. It has also been shown that some transposons have their ends consisting of insertion sequences (IS) which themselves resemble a transposon in having inverted repeats. This means that two identical transposons with an intervening sequence may give rise to a new but longer transposon (Fig. 15.7).

Fig. 15.6. Presence of inverted repeats at the two ends of each strand of a transposon in a plasmid and the formation of characteristic stem and loop in each strand on strand separation due to denaturation. Stem and loop formation is an evidence for inverted repeats.

Fig. 15.7. A plasmid with several transposons of varying sizes. Note the presence of a smaller transposon within a bigger transposon.











