Transposons in eukaryotes
On the basis of their structure, transposons in eukaryotes can be classified into four types. The basis of their classification includes the size and nature of the terminal repeats, which could be long or short and could be direct repeats (e.g. CCCAGAC and GGGTCTG) or inverted repeats (e.g. CCCAGAC and GTCTGGG). Following are the four types of transposons (i) with long terminal direct repeats (e.g. Copia in Drosophila, Ty in yeast and IAP in mice); (ii) with long terminal inverted repeats (e.g. FB, TE in Drosophila);
(iii) with short terminal inverted repeats (e.g. P and I in Drosophila; Ac/Ds in maize; Tam1 in snapdragon and Tc1 in Coenorhabditis)and (iv) without terminal repeats (Alu in mammals); In another classification, on the basis of mechanism of transposition, transposable elements are classified into two classes : (i) those, which transpose by reverse transcription of an RNA intermediate (DNA-RNA-DNA)—class I (e.g. copia like, Ty, IAP, LI, F, etc.) and (ii) those, which transpose directly, DNA to DNA—class II (e.g. P, Ac, Spm/En, Tam, Tc1).
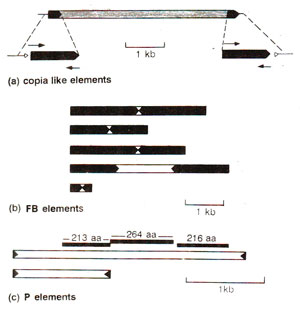
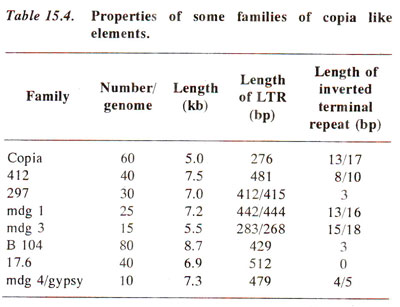
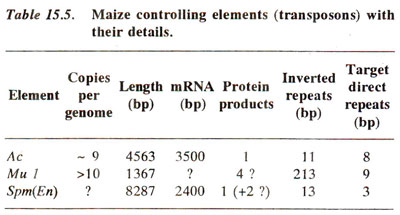
Transposable elements (TE) in Drosophila. As much as 10% of the genome in Drosophila may consist of transposons; most important of these are Copia like elements, the fold back (FB) and P (and I) elements. The salient features of these three types are shown in Figure 15.8.
(a) Copia like elements. The detailed features of Copia and Copia like elements are presented in Table 15.4, showing that their size may range from 5 to 8.7 kb and their number may range from 10 to 80. Each carries (i) a long direct terminal repeat (LTR) ranging from 268 to 512 bp and (ii) a short imperfect inverted repeat, ranging from zero to 18 bp. Insertion of these elements cause mutations, a classical example being white-apricot (wa)mutation for eye colour caused by insertion of a copia like element.
(b) FB elements. These elements range from a few hundred to a few thousand base pairs and carry long inverted repeats at the termini. The repeats may be adjoining (FB1, FB5) or may be separated (FB3, FB4, FB5, FB7, FB8, FB9), but can always fold back to form stem and loop structures. They may cause mutations by insertion or by their effect on gene expression (due to their presence near a control region). FB elements are also found flanking another class of transposons called TE (e.g. TE1, TE28, TE77, TE98). TE1 was first identified due to transposition of white eye locus from X-chromosome to second chromosome.
(c) P and I elements.The P and I elements are responsible for hybrid dysgenesis in Drosophila melanogaster, when strains like P, M, I and R are crossed in certain combinations. Hybrid dysgenesis is a phenomenon responsible for unusual characteristics like sterility, high mutation rate, high frequency of chromosomal aberrations and non-disjunction, which appears in the progeny, when either (i) M (maternal contributing) strain is crossed as female with P (paternal contributing) strain as male, or (ii) I (inducer) strain is crossed as male with R (reactive) strain as female.
These hybrid progeny are called dysgenic, hence the name hybrid dysgenesis. In these PM and IR systems, reciprocals (P as ♀ x M as ♂; I as ♀ x R as ♂)give normal progeny and produce no dysgenic progeny. It is also noted that while I-R interaction affects only females, P-M interaction affects both sexes in the progeny. The P and I strains contain P factors and I factors respectively. P factors are stable in P strain, but transpose when introduced in cytoplasm of M strain. Similarly, I factor is stable in I strain, but become unstable in R strain. P factor is absent in M strain and I factors is absent in R strain. Several mutations have also been identified to be due to hybrid dysgenesis. The P factor was found to be an element with 0.5-2.9 kb sequence and I factor with a 5.4 kb sequence. These may give rise to smaller-elements or factors (0.5 kb-1.4 kb) due to internal deletions. The P factor (2.9 kb) is flanked by inverted repeats of 31bp but no homology was found at the ends of I factor suggesting that no direct or inverted repeats are found at the ends of I factor.


 The current explanation of hybrid dysgenesis assumes synthesis of transposase and P-repressor by P element, the former causing transposition and the latter blocking transposition (by preventing transposase production). In P cytotype, high copy number of P gives high level of repressor leading to lack of transposition. However, in cross M♀ x P ♂, the zygote derives its cytoplasm from M having no repressor, leading to free transposition and damage throughout the genome, manifested as hybrid dysgenesis (Fig. 15.9).
The current explanation of hybrid dysgenesis assumes synthesis of transposase and P-repressor by P element, the former causing transposition and the latter blocking transposition (by preventing transposase production). In P cytotype, high copy number of P gives high level of repressor leading to lack of transposition. However, in cross M♀ x P ♂, the zygote derives its cytoplasm from M having no repressor, leading to free transposition and damage throughout the genome, manifested as hybrid dysgenesis (Fig. 15.9).
Ty elements in yeast. An example of Ty element in yeast is Tyl (Fig. 15.10), which is present in 35 copies in yeast genome. At the ends are present direct repeats (not inverted repeats or IR as found in bacteria) called delta (6), of which 100 copies per genome are found. Ty elements generate a repeated sequence (5bp long of the target DNA) during transposition and cause mutations due to insertion. Transposition in this case is caused through an RNA intermediate (class I transposons), Tam 1 element in snapdragon (Antirrhinum majus).In Antirrhinum majus, Tam 1 is responsible for pattern of colour seen on petals of some variegated strains and is 17 kb long. Taml is found in an unstable mutant, Nivea recurrens, where Tam 1 is present in the region of chalcone synthase gene and causes 3bp target site duplication. Tam 1 also has 13bp inverted repeats at its termini, which are identical to those in Spml8 of maize. Other transposable elements of this series are numbered Tam 2-Tam 10. Putative transposable elements are also suggested to be present in soybean.

Controlling elements in corn. The transposable elements, although studied initially only in bacterial cells, but were originally discovered in maize plant by Marcus Rhoades and Barbara McClintock. These genetic elements in maize were found to be responsible for turning the expression of genes on or off hence the name 'controlling elements'. It was also shown that these controlling elements when located on maize chromosomes represented specific sites for breakage and reunion of chromosomes leading to gross changes in chromosome structure.
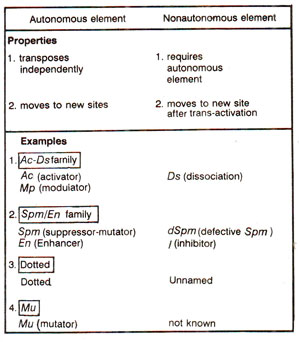
Several families of controlling elements or transposable elements have been isolated and studied in detail. Some of these are one element system (e.g. Dt or Dotted, Mp or modulator, Uq or ubiquitous, Bg or Bergamo, Mu or mutator, etc.) and others are two elements systems (e.g. Ac-Ds system, Spm-dSpm system, etc.).
It is possible that one element system also turn out to be two elements system on further investigations. The number, type and location of these elements are characteristic of each maize strain. In each two elements system, the elements can be classified into two classes : (i) Autonomous elements, which have the ability to excise and transpose, rendering sites of their insertion mutable or m alleles. (ii) Non-autonomous elements, which do not transpose and are unstable and can be derived from autonomous elements of the same family. They become unstable only when an autonomous member of the same family is present elsewhere in the genome.
When complemented by an autonomous member of the same family, the non-autonomous member may display properties of the autonomous element including its ability to transpose (Fig. 15.11; Table 15.5) A non-autonomous element of one family, can not be cross-inactivated by the autonomous element of any other family, so that the relationship between autonomous and non-autonomous elements of a family seems to be quite specific.

The details of autonomous elements of three different families of controlling elements in maize are presented in Table 15.5. These details demonstrate that these controlling elements have the same organization as bacterial transposons (see later).
In a two element system, a family consists of a solitary autonomous element associated with a variety of non-autonomous elements. These elements are characterized by inverted repeats (IR) at the ends and short direct repeats in the adjoining target DNA, as shown for other transposons (Table 15.5).
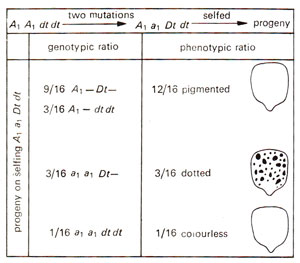
 (a) Dotted (Dt) element. Dotted (Dt)element was discovered by Marcus Rhoades in 1938, when from a ear of a pure breeding pigmented genotype of maize, among kernels a dihybrid segregation ratio of 12 : 3 : 1 (pigmented : dotted : colorless) was obtained. This was surprising Because no segregation was expected. The genetic analysis suggested that the original phenotype was controlled by two genes with the genotype A1 A1 dt dt and that two mutations (A1 → a1 and dt → Dt)gave the genotype A1 a1 Dt dt which gave the following phenotypic and abbreviated genotypic ratio (Fig. 15.12) : 12/16 pigmented (A1 Dt = 9/16 +A1dt = 3/16) : 3/16 dotted (a1 Dt): 1/16 colourless (a1 a1 dt dt).
(a) Dotted (Dt) element. Dotted (Dt)element was discovered by Marcus Rhoades in 1938, when from a ear of a pure breeding pigmented genotype of maize, among kernels a dihybrid segregation ratio of 12 : 3 : 1 (pigmented : dotted : colorless) was obtained. This was surprising Because no segregation was expected. The genetic analysis suggested that the original phenotype was controlled by two genes with the genotype A1 A1 dt dt and that two mutations (A1 → a1 and dt → Dt)gave the genotype A1 a1 Dt dt which gave the following phenotypic and abbreviated genotypic ratio (Fig. 15.12) : 12/16 pigmented (A1 Dt = 9/16 +A1dt = 3/16) : 3/16 dotted (a1 Dt): 1/16 colourless (a1 a1 dt dt).

The dotted phenotype (appeared in kernels as well as in leaves) in a1 Dt, would probably result from the reverse mutation (a1 → A1) in somatic cells, and the frequency of this mutation should be unusually high. This led to the discovery of a1 as an unstable mutant allele (its instability dependent on Dt)showing high rate of reverse mutation. It was also shown that once reverse mutations have occurred, they are stable, reverted A1 being transmitted to the progeny in a stable form. It means Dt causes mutations in a1 to A1, which remains stable.
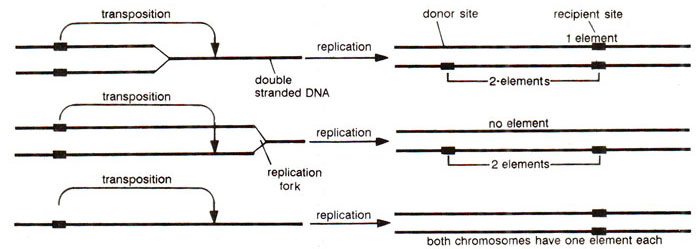
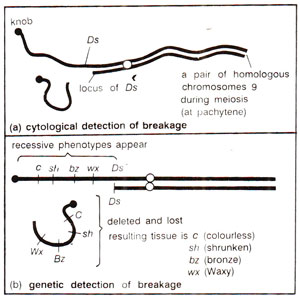
(b) Ac-Ds (activator-dissociation) system. Ac-Ds system is the most popular system and was discovered in 1950 by Barbara McClintock, when she found that the presence of a genetic factor Ds caused high tendency towards chromosome breakage at the location, where it appears. These breaks could be located either cytologically (at pachytene) or genetically by uncovering of recessive alleles in a heterozygote (Cc Shsh Bzbz Wxwx →homozygous recessive phenotype; Fig. 15.13). This property of Ds was dependent on another unlinked genetic factor Ac (activator) in the same manner as instability of a1was dependent on Dt. Thus, in presence of Ac, Ds may undergo transposition or may cause chromosome breaks and an increase in the number of Ac element in the genome decreases the frequency of transposition or chromosome breaks by Ds.

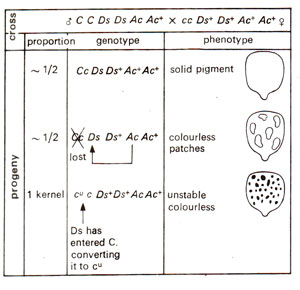
When McClintock tried to map Ac, she could not map it, because different plants showed its position on different chromosomes or on different positions of same chromosome. Ds was also found changing positions on an arm of chromosome 9, as shown by the uncovering of different gene combinations in different variegated regions. This suggested that Ds keeps on jumping positions, a property which was dependent on the presence of Ac. If Ds+ and Ac+ mean absence of these elements, the results of the following cross cc DsDs Ac+Ac+ (♀) x QC DsDs AcAc+ (♂) would further explain the behaviour of Ds and Ac elements. Both the possible genotypes in the progeny (Cc DsDs Ac+Ac+ and Cc DsDs AcAc+) should give coloured phenotype in kernels, but following two types of kernels were obtained, which helped in understanding further the attributes of Ac and Ds elements : (i) About 50% kernels had colourless patches, which were attributed to loss of C allele due to induced deletion, (ii) A single kernel was unstable colourless,
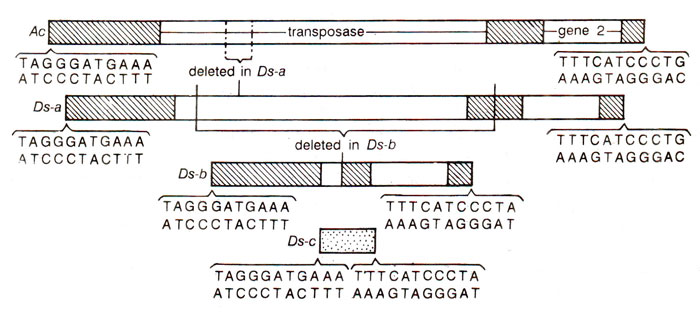
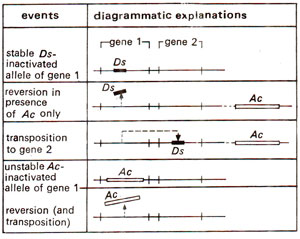
where Ds got inserted into C and inactivated it, producing colourless phenotype, which was unstable (cu) so that patches of colour appeared in the kernel. If Ac is removed from this line by crossing, cu now becomes a stable mutant (Fig. 15.14). The wx (waxy) and sh (shrunken) loci have been similarly affected by Ds in presence of Ac. A number of different mutable waxy alleles could also be produced due to insertion of Ds at different positions within the locus wx. In addition to Ds which causes mutability in presence of Ac, the latter (Ac)may also get inserted into the target gene, making it unstable. This inserted Ac may become Ds (non-autonomaus) spontaneously due to deletions or inactivation of the gene transposase, associated with it. The behaviour of Ac and Ds elements in corn is summarized in Figure 15.15.


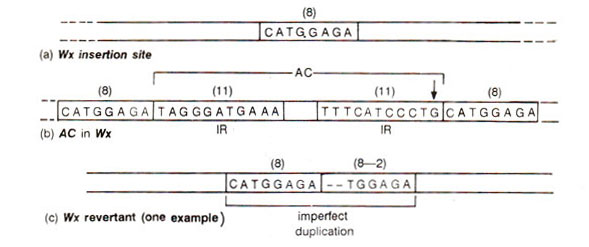
Molecular studies explained the above results in a very satisfactory manner, using Wx locus, whichwas rendered unstable by insertion of either Ac or Ds. Waxy mRNA was isolated from endosperm and cDNA clones were prepared from this mRNA. This cDNA was used for isolation of (i) Wx gene, (ii) wx gene containing either Ac or Ds and (iii) Wx revertants derived from unstable alleles. The isolated genes were sequenced and compared giving information about Ac (Table 15.5) and Ds (Fig. 15.16). It can be seen that Ds elements are Ac elements with deletions, which can be small (Ds-a)or large (Ds-b, Ds-c). Flanking the Ac or Ds element were also found 8bp long duplications of a sequence of Wx gene (Fig. 15.17). The mechanism of the origin of these duplications is shown in Figure 15.18.
It has been shown that transposition is caused by the enzyme transposase which acts on terminal inserted repeats (IR). In Ds, transposase gene being inactivated (due to deletions) but IR being present, the transposase synthesized by Ac, causes transposition, but as the number of Ac increases, repressor activity (self regulation; transposase may repress its own synthesis) may inhibit transposase synthesis leading to a decrease in frequency of transposition.
TheDs element was originally identified by its ability to provide a site for chromosome breakage upon activation by Ac. This breakage not only causes loss of alleles due to loss of an acentric fragment, but may also initiate a breakage-fusion-bridge cycle, that may lead to the appearance of sectors (due to loss of one dominant gene) and subsectors within sectors (due to further Joss of another dominant gene and so on).
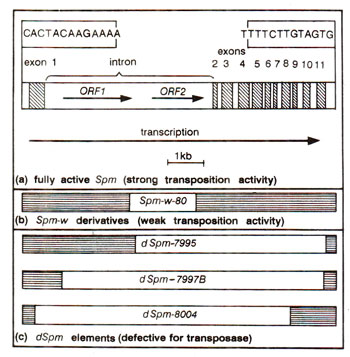
(c) Spm (suppressor-mutator) system.The suppressor-mutator family consists of an autonomous transposition competent Spm element (also designated as enhancer or En)and mutant element (e.g. dSpm = defective Spm), which can not transpose (Fig. 15.19). Suppressor mutator has dual properties : suppressor means ability to inhibit expression of gene at the locus occupied and mutator means ability to revert (transpose), so that the gene regains full activity. Sp+m- mutants have lost one, but not the other property.
The standard Spm (Spm-s)or En element is 8.3 kilobase (kb) long with 13 base-pair (bp) terminal inverted repetitions. It encodes a transposase enzyme and a positive autoregulatory gene product. The element has been used to produce transgenic tobacco plants, where Spm element transposes and trans activates dSpm excision in tobacco.
Transposable elements (TE) in Drosophila. As much as 10% of the genome in Drosophila may consist of transposons; most important of these are Copia like elements, the fold back (FB) and P (and I) elements. The salient features of these three types are shown in Figure 15.8.
(c) P and I elements.The P and I elements are responsible for hybrid dysgenesis in Drosophila melanogaster, when strains like P, M, I and R are crossed in certain combinations. Hybrid dysgenesis is a phenomenon responsible for unusual characteristics like sterility, high mutation rate, high frequency of chromosomal aberrations and non-disjunction, which appears in the progeny, when either (i) M (maternal contributing) strain is crossed as female with P (paternal contributing) strain as male, or (ii) I (inducer) strain is crossed as male with R (reactive) strain as female.

Fig. 15.9. The phenomenon of hybrid dysgenesis (due to insertion of P element) in Drosophila.

Fig. 15.10. The structure of yeast transposable elements Ty1 showing two copies of delta (5) sequence in direct orientation at each end.

Fig. 15.11. Relationship between autonomous and non-autonomous members of four families of controlling elements in corn.
Ty elements in yeast. An example of Ty element in yeast is Tyl (Fig. 15.10), which is present in 35 copies in yeast genome. At the ends are present direct repeats (not inverted repeats or IR as found in bacteria) called delta (6), of which 100 copies per genome are found. Ty elements generate a repeated sequence (5bp long of the target DNA) during transposition and cause mutations due to insertion. Transposition in this case is caused through an RNA intermediate (class I transposons), Tam 1 element in snapdragon (Antirrhinum majus).In Antirrhinum majus, Tam 1 is responsible for pattern of colour seen on petals of some variegated strains and is 17 kb long. Taml is found in an unstable mutant, Nivea recurrens, where Tam 1 is present in the region of chalcone synthase gene and causes 3bp target site duplication. Tam 1 also has 13bp inverted repeats at its termini, which are identical to those in Spml8 of maize. Other transposable elements of this series are numbered Tam 2-Tam 10. Putative transposable elements are also suggested to be present in soybean.

Fig. 15.10. The structure of yeast transposable elements Ty1 showing two copies of delta (5) sequence in direct orientation at each end.
Several families of controlling elements or transposable elements have been isolated and studied in detail. Some of these are one element system (e.g. Dt or Dotted, Mp or modulator, Uq or ubiquitous, Bg or Bergamo, Mu or mutator, etc.) and others are two elements systems (e.g. Ac-Ds system, Spm-dSpm system, etc.).

Fig. 15.11. Relationship between autonomous and non-autonomous members of four families of controlling elements in corn.
The details of autonomous elements of three different families of controlling elements in maize are presented in Table 15.5. These details demonstrate that these controlling elements have the same organization as bacterial transposons (see later).

Fig. 15.12. Appearance of dotted phenotype in corn, due to two simultaneous mutations
(A1→a1; dt→Dt).
(A1→a1; dt→Dt).

Fig. 15.13. Detection of breakage due to action of Ds element, (a) cytological level, (b) genetic level.

Fig. 15.14. Action of Ac-Ds system on endosperm colour (due to C) in corn (C = colour; c = colourless ; cu = unstable).

Fig. 15.15. A summary of the effects of Ac-Ds elements in corn, using two hypothetical genes.

Fig. 15.12. Appearance of dotted phenotype in corn, due to two simultaneous mutations
(A1→a1; dt→Dt).
(A1→a1; dt→Dt).
The dotted phenotype (appeared in kernels as well as in leaves) in a1 Dt, would probably result from the reverse mutation (a1 → A1) in somatic cells, and the frequency of this mutation should be unusually high. This led to the discovery of a1 as an unstable mutant allele (its instability dependent on Dt)showing high rate of reverse mutation. It was also shown that once reverse mutations have occurred, they are stable, reverted A1 being transmitted to the progeny in a stable form. It means Dt causes mutations in a1 to A1, which remains stable.
(b) Ac-Ds (activator-dissociation) system. Ac-Ds system is the most popular system and was discovered in 1950 by Barbara McClintock, when she found that the presence of a genetic factor Ds caused high tendency towards chromosome breakage at the location, where it appears. These breaks could be located either cytologically (at pachytene) or genetically by uncovering of recessive alleles in a heterozygote (Cc Shsh Bzbz Wxwx →homozygous recessive phenotype; Fig. 15.13). This property of Ds was dependent on another unlinked genetic factor Ac (activator) in the same manner as instability of a1was dependent on Dt. Thus, in presence of Ac, Ds may undergo transposition or may cause chromosome breaks and an increase in the number of Ac element in the genome decreases the frequency of transposition or chromosome breaks by Ds.

Fig. 15.13. Detection of breakage due to action of Ds element, (a) cytological level, (b) genetic level.

Fig. 15.14. Action of Ac-Ds system on endosperm colour (due to C) in corn (C = colour; c = colourless ; cu = unstable).

Fig. 15.15. A summary of the effects of Ac-Ds elements in corn, using two hypothetical genes.
TheDs element was originally identified by its ability to provide a site for chromosome breakage upon activation by Ac. This breakage not only causes loss of alleles due to loss of an acentric fragment, but may also initiate a breakage-fusion-bridge cycle, that may lead to the appearance of sectors (due to loss of one dominant gene) and subsectors within sectors (due to further Joss of another dominant gene and so on).
(c) Spm (suppressor-mutator) system.The suppressor-mutator family consists of an autonomous transposition competent Spm element (also designated as enhancer or En)and mutant element (e.g. dSpm = defective Spm), which can not transpose (Fig. 15.19). Suppressor mutator has dual properties : suppressor means ability to inhibit expression of gene at the locus occupied and mutator means ability to revert (transpose), so that the gene regains full activity. Sp+m- mutants have lost one, but not the other property.