Pribnow box and other sequences common to DNA regions upstream to several operons
Upstream to starting point of lac operon are number of sequences which are common to many E. coli and bacteriophage genes. There is a sequence at about 10 bases upstream from starting point (also called minus ten or - 10 sequence) called Pribnow box which is an important sequence and named after D. Pribnow, who first discovered it in 1975.
Pribnow box is shown in Figure 35.21 and has the following sequence in lac operon.
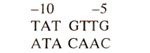
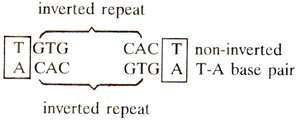
This CAP binding site is common to lac,gal and ara operons. It lies within -53 to -23 bp in gal operon,bctween -72 to -52bp in lac operon and between -107 to -78bp in ara operon.




