Resistance to antibacterial agents
Resistance occurs when a previously susceptible organism is no longer inhibited by an antibiotic at a concentration that can be safely achieved in clinical practice. Resistance can develop quickly because:- bacteria multiply rapidly;
- mutations arise regularly;
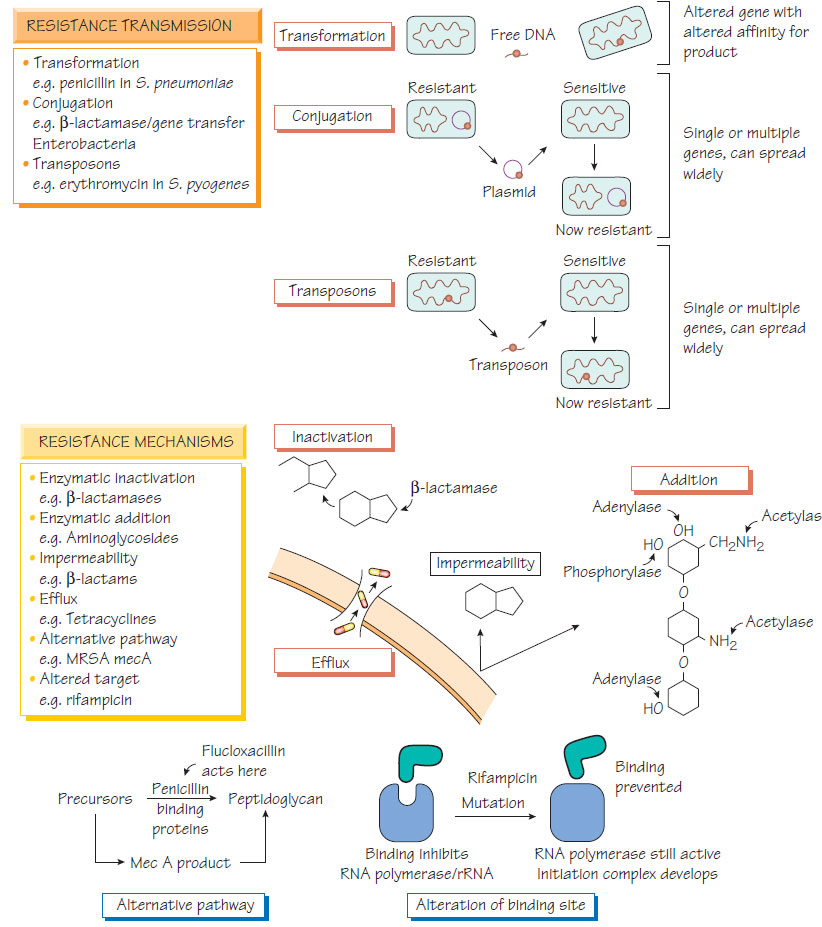
- segments of DNA can transfer by transformation;
- genes can be transferred rapidly by bacteriophages, plasmids or other mobile genetic elements.
Antibiotic use favours the survival of resistant organisms. The replication of organisms that have accidentally developed mechanisms to avoid destruction can pose a threat to the successful treatment of infections.
Transformation
Many bacterial species incorporate naked DNA into their genome, a process called transformation. For example, Streptococcus pneumoniae and Neisseria gonorrhoeae incorporate small sections of penicillin-binding protein genes from closely related species to produce a penicillin-binding protein that binds penicillin less avidly, so becoming more resistant. Such organisms are still able to synthesize peptidoglycan and maintain their cell walls in the presence of penicillin.
Conjugation
Bacteria contain plasmids, circular DNA structures that are found in the cytoplasm. Many genes are carried on plasmids including those that encode metabolic enzymes, virulence determinants and antibiotic resistance. Plasmids can pass from one bacterium to another by conjugation allowing 'resistance genes' to spread rapidly in populations of bacterial species that share the same environment (e.g. within the intestine). Combined with antibiotic selective pressures (e.g. in hospitals) that favour the survival of organisms with resistance plasmids, multiresistant populations may develop.
Transposons and integrons are mobile genetic elements able to encode transposition and move between the chromosome and plasmids, and between bacteria. Many functions, including antibiotic resistance, can be encoded on a transposon. Resistance to methicillin among Staphylococcus aureus and to tetracycline among N. gonorrhoeae probably entered the species by this route. Integrons are important in transmission of multiple drug resistance in Gram-negative pathogens. Resistance genes can also be mobilized by bacteriophages (viruses that live in bacteria).
Multiple resistance
Multiple resistance can develop on mobile genetic elements because once a gene is established on the element, it can readily acquire resistance to another agent by one of the mechanisms above. Once there is more than one resistance gene, exposure to any of these agents will permit survival of the organism, which increases the risk of further resistance being acquired.
Antibiotic modification
Enzyme inactivation
A common resistance mechanism is degradation of the antibiotic. Many strains of S. aureus produce an extracellular enzyme (β- lactamase), which can break open the penicillin β-lactam ring, thereby inactivating it. In the face of newer β-lactam antibiotics, many human pathogens have acquired a range of genes that encode broad-spectrum β-lactamases; these include Escherichia coli, Haemophilus influenzae and Pseudomonas spp. The genes are often found on mobile genetic elements (transposons). The spread of different types of extended-spectrum β-lactamases (ESBLs), such as CTX-M and AmpC, among Enterobacteriaceae is producing resistance to cephalosporins and broad-spectrum penicillins in organisms that cause hospital-associated infections. Spread to the community has already occurred.
Some bacteria express enzymes that add an inactivating chemical group to the antibiotic, so inhibiting its activity. Bacteria may become resistant to aminoglycosides by adding an acetyl, amino or adenosine group to the antibiotic molecule. Different aminoglycosides differ in their susceptibility to this modification, amikacin being the least susceptible. Aminoglycoside-resistance enzymes are found in Gram-positive organisms (e.g. S. aureus) and Gramnegative organisms (e.g. Pseudomonas spp).
Impermeability
Some bacteria are naturally resistant to antibiotics because their cell envelope is impermeable to that particular antibiotic (e.g. Pseudomonas spp. are impermeable to some β-lactam antibiotics). Aminoglycosides enter bacteria by an oxygen-dependent transport mechanism and so have little effect against anaerobic organisms. Other bacteria may lose a porin protein, so creating a permeability barrier that stops antibiotics from entering the cell.
Bacteria, for example E. coli or streptococci, may become resistant to tetracyclines, macrolides or fluoroquinolones by the acquisition of an inner membrane protein that actively pumps the antibiotic out of the cell - an efflux pump.
Alternative pathway
Bacteria may acquire genes that create an alternative pathway that can circumvent the metabolic block imposed by an antibiotic. S. aureus becomes resistant to methicillin or flucloxacillin when it acquires the gene mecA, which encodes an alternative penicillinbinding protein (PBP2') that is not inhibited by methicillin. Although the composition of its cell wall is altered, the organism is still able to multiply.
Rifampicin acts by inhibiting the β-subunit of RNA polymerase. Resistance develops when the RNA polymerase gene is altered by point mutations, insertions or deletions. The new RNA polymerase is not as easily inhibited by rifampicin and resistance occurs. Similarly, an alteration of the binding sites on DNA gyrase (the target of fluoroquinolones) can make an organism resistant. The genes responsible for these effects are often found in a small region of the target gene, for example in the rifampicin resistancedetermining region (RRDR).