Ion Transport
Loss- or gain-of-function experimentation has identified many determinants that
control and mediate Na
+ and K
+ uptake, as well as homeostasis
in planta (Amtmann and Sanders, 1999; Hasegawa
et al., 2000b; Horie and Schroeder,
2004; Maathuis
et al., 1996; Mäser
et al., 2002; Niu
et al., 1995; Schachtman, 2000;
Sondergaard
et al., 2004; Tester and Davenport, 2003; Zhu, 2003). Illustrated in
Fig. 12.1A are known or suspected cellular Na
+ uptake systems, as well as other
facilitators of homeostasis such as aquaporins and the proton transporters that
establish membrane potentials in different compartments (Gaxiola
et al., 2002).
Included are channels and transporters responsible for K
+ and Na
+ flux and H
+ pumps that generate the requisite electrochemical potential necessary to facilitate
channel function or secondary-active transport (Arango
et al., 2003; Borsani
et al., 2001; Horie and Schroeder, 2004; Schachtman, 2000; Tester and Davenport, 2003;
Vitart
et al., 2001; Ward
et al., 2003; Zhu, 2003). Other,
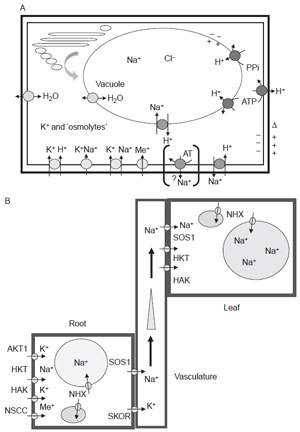 |
| FIGURE 12.1 Transporters in plant cells and
transport of sodium in plants. (A) Complexity of
transporters and facilitators involved in ion
homeostasis. A cellular view of ion and water
facilitators located in the plasma membrane or
tonoplast membrane, although the location of a
number of these proteins in other membranes
(e.g., the prevacuolar complex, plastids,
mitochondria, or the endomembrane system) is
not excluded, and has rarely been investigated.
White symbols—proton ATPases and inorganic
pyrophosphatases; dark grey symbols—sodium/
proton antiporters (sos1 and NHXn); light grey
symbols—different K+ (or alkali ion) transporters
(HKT, KAT, HAK, CNGC, NSCC); stippled
symbol—aquaporins (and/or low molecular
weight
metabolite facilitators); dotted
symbols—the presence of yeast ENAx-like
(bracketed) sodium
ATPases has not been
observed in plants. However, the transfer of the
yeast ENA1 gene into
tobacco suspension culture
cells resulting in altered salt stress tolerance has
been reported (Marin et al., 2003).
(B) Long-distance K+ and Na+ transport.
The distribution of sodium in plants is
controlled
by a limited number of potassium (or alkali ion)
transporters and sodium/proton
antiporters
(Tester and Davenport, 2003). The transport of
sodium and redistribution into sinks,
such as
specialized cells, inert tissues, or vacuoles,
generates a gradient that can be maintained as
long as spaces are available into which sodium
can be caged, and as long as water movement
can
be maintained. In at least some halophytes,
the gradient can have a reversed orientation, with
sodium accumulation in distal plant parts, thus
generating an osmotic potential that draws water
into aerial tissues. |
yet unidentified, genetic loci
are involved in the control of Na
+ and K
+ homeostasis, including those that
regulate Ca
2+ homeostasis (Nublat
et al., 2001; Rus
et al., 2001). Focal are Na
+ transport systems that control not only intracellular distribution of Na
+ but also
homeostasis between tissues and organs (Fig. 12.1B) (Berthomieu
et al., 2003;
Horie and Schroeder, 2004; Laurie
et al., 2002; Rus
et al., 2004; Tester and
Davenport, 2003; Ward
et al., 2003; Yokoi
et al., 2002). Possibly most important is
the salt overly sensitive (
sos) pathway described from
Arabidopsis, described in
detail below (Aharon
et al., 2003; Gaxiola
et al., 2001; Quintero
et al., 2002; Su
et al., 2001; Talke
et al., 2003). One of the three proteins in this pathway,
sos1,
controls Na
+ uptake into the root xylem. It is hypothesized that high-affinity
K
+ transporter 1 (HKT1), originally described as a K
+ transporter, acts as a
Na
+ transporter that recirculates the ion from the shoot to the root when Na
+ is
in excess (Horie and Schroeder, 2004; Munns, 2002; Rus
et al., 2004; Shi
et al., 2003;
Yokoi
et al., 2002). Phenotypic suppression of
sos1–1 NaCl sensitivity by dysfunctional
hkt1 alleles is genetic evidence that these two transport systems have
opposing functions in Na
+ homeostasis (Laurie
et al., 2002). Thus,
sos1 and
HKT1 may function in concert to regulate Na
+ content in the shoot. However,
AtHKT1 overexpression does not increase salt tolerance of
Arabidopsis as would be expected (Laurie
et al., 2002). Most likely the endomembrane cation/H
+ transporters,
exemplified by the NHX family, are principally responsible for vacuolar
compartmentalization of Na
+ in plant cells (Aharon
et al., 2003; Munns, 2002;
Rigas
et al., 2001).
However, NHX gene copy number and their expression under
nonsaline conditions might indicate that the various NHX forms could have very
different, additional functions or that their function under saline conditions
becomes modified.
Na
+ detrimentally affects K
+ acquisition. K
+ is an essential macronutrient that
functions in critical processes ranging from charge balance, osmotic adjustment,
and enzyme catalysis to growth and development (Elumalai
et al., 2002; Maathuis
et al., 1996; Rains and Epstein, 1965). Na
+ disturbs intracellular K
+ homeostasis
possibly because it can compete for binding sites on enzymes and transport
proteins (Cramer
et al., 1987; Epstein, 1961; Niu
et al., 1995; Tester and Davenport,
2003). Ca
2+ enhances K
+ /Na
+ selective accumulation because it facilitates K
+ uptake (Epstein, 1961; Hasegawa
et al., 2000b; Li
et al., 1998).





