The Context of Salinity Stress
The considerable increase in worldwide crop production that occurred during the
green revolution did not result in substantially greater land use but focused on
adapting germplasms to respond to altered farm management practices
(Trewavas, 2001). However, even with better adjustment of crops and increased
production efficiency, the actual yield is less than the crop genetic potential.
By now, the increased population in developing countries puts even more constraints
on production as urban populations compete with agriculture for fresh
water. In parts of the world, this has necessitated the use of less suitable irrigation water, often water of low quality that is unsuitable for high-yield agriculture with existing crops. This in turn makes it paramount to find ways that realize the
genetic potential and yield capacity of crop genomes, even under moderate
stresses, or to enhance them by transgenic means.
Significant among the abiotic stresses is salinity, which not only constraints
crop production in a particular growing cycle but also leads to steady deterioration
of soils and irrigation water that compounds the effects of salinity on
subsequent crop generations. In many countries around the globe where water
is already scarce and droughts are recurring, soil salinity is a major constraint
to crop productivity that negatively affects much of the cultivated land and
substantially reduces yield (Flowers and Yeo, 1995; Läuchli and Epstein, 1990;
Maas, 1990; Munns, 1993). These facts, and the prospect of erratic rainfall patterns
that could increase in the future, have led to efforts to improve yield stability of
present-day crops by focusing on abiotic stress factors (Flowers, 2004; Flowers and
Yeo, 1995; Serrano, 1996). Crop improvement strategies seek to develop more saltadapted
or -adaptable germplasms by utilizing molecular genetic approaches and
resources developed during the mid-1980s, notably marker-assisted breeding
techniques, exploration of halophytic species, biotechnology, and genomics (Apse
and Blumwald, 2002; Garciadeblas
et al., 2003; Hasegawa
et al., 2000b;Koyama
et al., 2001; Loudet
et al., 2003; Ribaut and Hoisington, 1998; Tuberosa
et al., 2002;
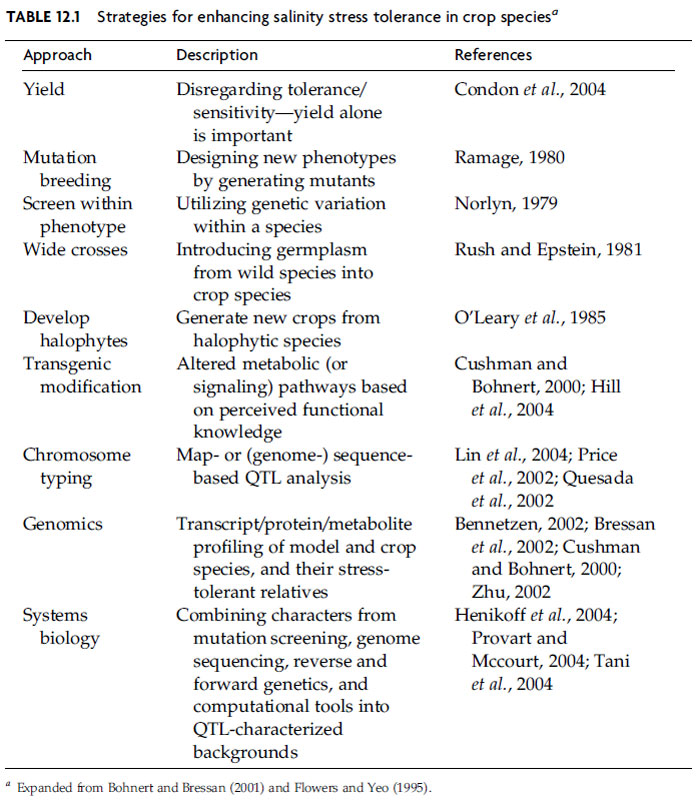
Zhu, 2002). Table 12.1 lists strategies that have been suggested, initially with
respect to different classical breeding approaches, and more recently, including
genome-anchored methods that can significantly enhance breeding because physical
maps, mapped and phenotyped mutants, and genome sequences provide
precision.
High salt in the root environment, salinity that typically appears in the form of
increased NaCl in the soil, is a combination of ionic and hyperosmotic imbalance
and secondary effects including pathologies that inhibit growth and can affect
development or cause cell death (Hasegawa
et al., 2000a; Zhu, 2001, 2002). Ionic
and osmotic stress signals are sensed and decoded by all plants via distinct and
interconnecting signal pathways that are response relays for the control of unique
and stress-specific programs. These pathways are response relays that control
genetic programs and coordinate determinants and processes required for adaptation.
Both stress conditions constitute environmental perturbations that modulate
normal cellular or developmental programs (Zhu, 2002, 2003). Consequently,
salt adaptation involves determinants that establish ion homeostasis and/or
osmolyte biosynthesis, termed osmotic adjustment. In cases where the severity
of a stress condition exceeds the capacity of a species, ecotype or line to acclimate,
this then precludes cell division, expansion and normal development and may
result in death (Apse and Blumwald, 2002; Blumwald, 2000; Hasegawa
et al., 2000b; Zhu, 2001, 2002).
The capacity of species to adapt to salt stress distinguishes
glycophytic species with a reduced capacity from halophytes that are to various
degrees able to adapt well, or even grow better at slightly increased levels of
sodium, and many of the latter may in fact use NaCl as a ‘‘cheap’’ osmoticum
(Adams
et al., 1998).
Evolutionary adaptations have resulted in species that exhibit different competence
to tolerate or resist high salt and complete their life cycles. Although
glycophytes and halophytes differ substantially in their capacity to tolerate salt,
the cytosolic and organellar machineries of the two plant categories seem to be
equally sensitive to Na
+ and Cl
- (Flowers, 2004; Greenway and Osmond, 1972;
Hasegawa
et al., 2000b; Jacoby, 1999; Serrano, 1996). Consequently, adaptation by
plants in both groups requires cellular responses that attenuate the osmotic and
ionic components of salt stress. The options are limited. They involve NaCl
exclusion or compartmentalizing Na
+ and Cl
- into an ‘‘inert’’ compartment, vacuole, or tissue. Other response mechanisms, such as avoidance reactions, in
essence induced dormancy, are not considered here. Simultaneously, mechanisms
that confine salt must be accompanied by the accumulation of solutes that are
compatible with cellular metabolism. Such osmolytes must increase in cytosol and
organelles to achieve osmotic adjustment (Blumwald, 2000; Hasegawa
et al., 2000a,b; Zhu, 2002, 2003). Both osmotic adjustment and the confinement of
sodium in (pre)vacuoles have been shown in the single cell model,
Saccharomyces cerevisiae (Gaxiola
et al., 1999; Hohmann, 2002).
That is, the mechanisms by which
all plants achieve osmotic and ionic equilibria are mediated by orthologous
mechanisms based on conserved biochemical and/or physiological functions
that are inherently necessary for essential plant processes (Hasegawa
et al., 2000a; Serrano
et al., 1999; Van Camp, 2005; Van Camp
et al., 1996; Zhu, 2000,
2001). This statement has been substantiated by the genomic DNA sequences of
two glycophytes,
Arabidopsis thaliana and
Oryza sativa, which seem to include all
components that have been researched as essential or necessary for plants to cope
with salt stress in different model species and crops (
Arabidopsis Genome
Initiative, 2000; Goff
et al., 2002; Yu
et al., 2002).
What then, if the important stress tolerance components are ubiquitous,
distinguishes glycophytes and halophytes? To solve this conundrum, research is
directed into several areas. One is to determine if halophytic versions of salt
adaptation determinants have greater innate operational capacity to facilitate
survival, growth, and development in saline environments, that is, if halophytic
versions of genes may represent an allele that encodes a more effective protein
that functions in the presence of high salt (Waditee
et al., 2002). An example
supporting such a view may be the case of L-
myo-inositol-1-phosphate synthase
that distinguishes rice (
O. sativa) from a wild relative (
Porteresia coarctata).
In
Porteresia, the homodimeric enzyme retains its aggregation state in high salt,
while the rice protein disintegrates into enzymatically inactive monomers at
much lower salt concentrations. This may be due to a domain that discriminate
the two forms of the enzyme, and, indeed, overexpression of the
Porteresia enzyme
enhances salt tolerance (Majee
et al., 2004).
Alternatively, halophytes may control universal determinants in a manner that
imparts to the species a preadapted state or a faster and superior ‘‘adaptive
response capacity’’ when the saline environment becomes increasingly severe.
A point in favor of such a scenario may be studies targeting the
Arabidopsis relative
Thellungiella halophila (salt cress), which is salt tolerant. Preliminary transcript
profiling and analysis of expressed sequence tags (ESTs) seems to indicate that
the salt cress constitutively shows high nonstress activities for a range of genes/
transcripts, and that induction of these transcripts is initiated at a higher stress
level than in
Arabidopsis (Inan
et al., 2004; Taji
et al., 2004). The fact that (eu)
halophytes show increased growth at moderate concentrations of NaCl, higher
than in fresh water, might be causally related to the high constitutive expression of
stress response pathways.
Third, outlining a related hypothesis, it must also be considered that some
halophytes have evolved specialized adaptations (e.g., salt glands for excretion or
bladder cells for the storage of NaCl). It is therefore possible that such species also possess other unique determinants with specialized function to mediate adaptation
that are missing from the genomes of glycophytes. Such uniqueness will only
be revealed when we have identified the relevant genes in halophytic models.
A variation of this theme is chromosomal context and genome size. The fusion of
genomes during speciation or endoreduplication events that have been documented
for many species in diverse plant families may have resulted in duplicated
genes, paralogues of ubiquitous genes that further evolved in some species in
response to changing environments (Arango
et al., 2003). Such genes could,
although they might not be superior in their biochemical function to those in
glycophytes, impart higher tolerance to a species by their expression at constitutively
higher levels, in a stress-inducible manner, in different compartments, or by
being connected to altered or novel regulatory circuits. We will briefly review salt
stress tolerance mechanisms and transgenic approaches that have begun to engineer
ionic and osmotic tolerance mechanisms into model species. Subsequently,
we will place emphasis on the regulatory circuits that control mechanisms of
tolerance acquisition (Schachtman, 2000; Zhu, 2002).