Aminoacyl–tRNA Synthetases
Decoding of the protein message occurs at the ribosome, after prior attachment of amino acids to the tRNA molecules. Aminoacyl–tRNA synthetases (AARSs) are the family of enzymes responsible for covalent attachment of each amino acid to its correct, or cognate, tRNA molecule. This first step in protein synthesis is responsible for establishing the rules of the genetic code (which codon corresponds to which amino acid), as the aminoacylated tRNA has both the nucleic acid component of the genetic message (the anticodon) and the amino acid. In most organisms, there are 20 AARSs, one for each amino acid. Because of codon degeneracy, there is at least one tRNA per amino acid; serine, for example, can be attached to one of five different isoaccepting tRNA molecules. Although each enzyme catalyzes the same aminoacylation reaction, the substrates are unique and must be effectively selected from among the cellular pool. For example, valyl– tRNA synthetase (ValRS) binds the amino acid valine specifically and disregards the other 19 amino acids; it also recognizes tRNAs containing the anticodons GUN (where N is any nucleotide) and only attaches valine to these molecules.A. Amino Acid Attachment
Covalent attachment of an amino acid to its cognate tRNA occurs in a two-step mechanism catalyzed by an AARS (E). In the first step, energy is consumed in the form of ATP to convert the enzyme-bound amino acid to its highenergy aminoacyl adenylate (AA–AMP). In the second step the activated amino acid is transferred to the 3´ end of the cognate tRNA, where an ester bond is formed between the carboxyl group of the amino acid and either the 2´- or 3´-hydroxyl on the terminal adenosine’s ribose sugar to yield AA–tRNA. While for most AARSs the first step is tRNA-independent, glutaminyl–, glutamyl–, and arginyl–tRNA synthetases require cognate tRNA binding for aminoacyl adenylate formation.
| E + AA + ATP |
B. Selection of tRNA
Because of the critical need for accurate tRNA aminoacylation in protein synthesis, AARSs must be highly discriminating in substrate selection. Structural studies revealed a large area of contact between AARS and tRNA, providing numerous opportunities for sequence-specific recognition, including contacts with the many modified nucleotides of tRNAs. The majority of identity determinants lie in the anticodon and the acceptor stem (near the site of amino acid attachment) of the tRNA molecule. Although the genetic code links amino acid identity to anticodon sequence, an “operational RNA code” exists withintheacceptor stems of tRNAs. This operationalRNA code is considered by some to be a “second genetic code.” It relates sequence and structure in the acceptor stem to aminoacylation with specific amino acids.
Because determinants for aminoacylation are found in the acceptor stem, RNA minihelices are substrates for aminoacylation by at least 10 different aminoacyl– tRNA synthetases. A striking example is alanyl–tRNA synthetase, which makes no contact with the tRNAAla anticodon. Instead, the enzyme recognizes a unique G3:U70 base pair in the acceptor stem of the tRNA. Substitutions at this location eliminate aminoacylation of the tRNAwith alanine, while introduction of this base pair into noncognate tRNAmolecules target them for alanylation. An RNA “minihelix” (Fig. 2) is also efficiently aminoacylated with alanine provided it contains the critical G3:U70 pair. Thus, selection of the cognate tRNA by an AARS is based on a limited number of sequence or structural elements, and in some cases does not involve the anticodon triplet of the genetic code.
C. Amino Acid Selection
Because of their small size and limited chemical diversity, selection of a cognate amino acid from the cellular pool presents a greater challenge to the AARSs than does selection of a specific tRNA. Differentiation of amino acids is primarily achieved through the formation of a binding pocket specific for the cognate amino acid within the active site of the enzyme. In thisway specific hydrogen bond contacts can be made to a polar amino acid, or a positively charged amino acid can fit into a negatively charged cleft within the AARS. Amino acids with hydrophobic side chains have to be discriminated on the basis of shape.
In some cases binding of cognate tRNA influences amino acid selection. Glutaminyl–tRNA synthetase (GlnRS) is one of three AARSs that require the presence of cognate tRNA for aminoacyl adenylate formation. Biochemical and genetic studies demonstrated that conserved residues of GlnRS that contact the 3´-terminus of tRNAGln are required to properly form the glutamine binding site. The presence of noncognate tRNAs decreased the enzyme’s affinity for glutamine because only tRNAGln was properly oriented to allow formation of the glutamine binding pocket.
D. Editing by AARSs
Binding interactions alone can not completely discriminate against all noncognate amino acids. Some enzymes misactivate noncognate amino acids at a significant frequency and require an additional accuracy mechanism. The most well-characterized example is isoleucyl–tRNA synthetase (IleRS), responsible for activating isoleucine and attaching it to tRNAIle at the exclusion of valine, which is smaller than isoleucine by a single methylene group. IleRS is more successful in discriminating against larger or bulkier amino acids, but the smaller valine simply fits more loosely in the enzyme active site.
Despite misactivating valine as frequently as once per 180 correct (isoleucine) activations, the enzyme is able to maintain the required fidelity of tRNA aminoacylation through an active editing function, so that valine is not misincorporated into proteins to a significant extent. This editing is twofold, corresponding to the two steps of the aminoacylation reaction: pretransfer editing by IleRS selectively hydrolyzes the noncognate valyl–adenylate, while post-transfer editing hydrolyzes any valyl–tRNAIle that is formed.
| IleRS:Val-AMP + tRNAIle → IleRS + Val + AMP + tRNAIle (Pretransfer) IleRS:Val-tRNAIle → IleRS + Val + tRNAIle (Post-transfer). |
Both activities are dependent on the presence of cognate tRNAIle, particularly the D-arm, as determined by studies that investigated the ability of mutants of tRNAIle to trigger editing. Furthermore, the editing site on IleRS has been located within a peptide (connective polypeptide 1, CP1) inserted between the two halves of the Rossmann nucleotide binding fold that makes up the catalytic domain of Class I AARSs (see following). Valyl–tRNA synthetase (ValRS) and leucyl–tRNA synthetase (LeuRS) have sequences in CP1 similar to IleRS; it is interesting to note that these enzymes all recognize amino acids with aliphatic side chains. Isolated CP1 domains from IleRS and ValRS have been shown to execute post-transfer editing.
The location of the editing site in IleRS has been visualized by X-ray crystallography and is approximately 25 Å from the “synthetic” active site. Apparently both the aminoacyl adenylate and the aminoacylated tRNA must be translocated from the synthetic active site to the editing active site for an accuracy check before being released. Translocation of the misaminoacylated tRNA to the editing site, and subsequent proofreading, has been proposed to be triggered by a conformational change in the tRNA. How the isolated aminoacyl adenylate is translocated remains unknown. The overall mechanism for accurate aminoacylation by IleRS has been termed a “double-sieve”—amino acids that don’t fit into the active site binding pocket are excluded based on size, while valine and smaller amino acids are actively hydrolyzed at the second (editing) site when they are misactivated or subsequently attached to tRNAIle.
E. Class Organization of AARSs
Detailed structural information from X-ray crystallographic studies is available for nearly all of the 20 AARSs. As more and more sequences and structures were determined the synthetases were seen to be partitioned into two classes of 10 enzymes each, based on similarities in their catalytic cores.
Enzymes belonging to Class I function as monomers or homodimers (α2). Their active sites contain a Rossmann nucleotide binding fold, also observed in dehydrogenases and other nucleotide-binding proteins. This fold consists of an alternating pattern of β-strands and α-helices. The nucleotide binding fold is split into two halves, with a variable-length polypeptide insertion known as CP1 between the halves. As described earlier, this polypeptide makes up the editing domain present in some enzymes. Comparison of Class I enzymes identified a signature sequence within the active site; this stretch of 11 amino acids is located in the first half of the nucleotide binding fold and ends in HIGH (the one-letter code for His–Ile–Gly–His). The second half of the fold contains another conserved sequence, KMSKS, which also makes critical contributions to the enzyme active site.
Class II AARSs were first distinguished by their lack of the Class I signature sequence, but this subfamily also has distinct features of its own. They are mostly α2 dimers, although some are α2β2 tetramers. The enzymes belonging to Class II have active sites composed of a seven-stranded antiparallel β-sheet with three α-helices. The characteristic Class II sequence motifs 1, 2, and 3 exhibit only minimal sequence conservation, but contribute a helix-loopstrand, strand-loop-strand, and strand-helix to the active site, respectively.
A mechanistic difference between the enzymes of Classes I and II is the site of initial amino acid attachment to the tRNA. Class I enzymes aminoacylate the 2´-hydroxyl of the terminal adenosine’s ribose and most Class II enzymes use the 3´-hydroxyl. The amino acid subsequently migrates between the two positions. This early functional observation was rationalized once high-resolution enzyme–tRNA cocrystal structures were determined. Class I enzymes approach the tRNA acceptor stem from the minor-groove side of the RNA helix and are nearer the ribose 2´-hydroxyl, while Class II enzymes approach from the major-groove side of the tRNA, near the 3´-hydroxyl.
F. Mechanistic Clues from Structural Studies
Enzyme–tRNA cocrystal structures have also provided clues to the catalytic mechanisms used by AARSs. In the case of the Class I glutaminyl-tRNA synthetase (GlnRS) complexed with its cognate tRNAGln, the structure of the complex identified variations in the tRNA architecture compared with uncomplexed tRNA. GlnRS makes essential contacts with nucleotides in the tRNAGln anticodon, and in the cocrystal structure these nucleotides were indeed splayed out from their normal positions to interact closely with amino acids of the enzyme. Furthermore, the acceptor end of the tRNA showed significant distortion compared to the expected helical structure. The first base pair of the acceptor helix was broken, and the 3´-end of the tRNA bent back in a hairpin conformation toward the rest of the helix (Fig. 3). Such a folded-back orientation of the acceptor end of tRNAGln is necessary for the 3´-terminus to reach the glutaminyl adenylate in the enzyme active site. Furthermore, amino acid residues in the enzyme’s active site are inserted into the acceptor stem nucleotides to break the first base pair and facilitate the distortion of the end of the helix. Binding of the cognate anticodon may trigger this distortion, which is necessary for efficient transfer of the activated amino acid to the 2´-OH of the bound tRNA. Because the anticodon binding site is located a significant distance from the enzyme active site, tRNA recognition in this case has elements of a signal transduction mechanism.
G. Origins of Aminoacylation
As discussed earlier, AARSs have core catalytic domains that perform the functions of aminoacyl adenylate formation and transfer of the amino acid to the cognate tRNA. The sequences and structures of these domains also differentiate the enzymes as belonging to Class I or II. In addition to this class-defining active site domain, most AARSs also have one or more appended domains that are unique. These idiosyncratic domains often make specific contacts with recognition elements outside the tRNA acceptor stem, for example, at the anticodon or variable loop of the tRNA molecule (Fig. 4). In addition to the twodomain (or more) organization of the AARS enzymes, tRNAs can also be viewed as modular structures. As mentioned earlier, the acceptor stem and TΨC arm coaxially stack to form one portion of the L-shaped tRNA structure, while the D and anticodon arms stack to make the other tRNA arm (Fig. 2). The acceptor arm makes contacts with the catalytic core of the enzyme and contains the amino acid attachment site, while the anticodon, located on the second arm of the tRNA, is recognized by an appended domain.
Given this apparent segregation of functions between both AARS enzyme and tRNA substrate, it is interesting to consider whether an early protein synthesizing system might have used minimalist versions of both tRNA and AARS. Such a minimalist system is suggested by the observation that small RNA substrates based on the acceptor stem sequence but lacking the anticodon (such as the minihelices discussed earlier) are substrates for many AARSs. In the case of the tRNA substrate, a minihelix containing the amino acid attachment site would have been the earlier part of the tRNA, while the anticodon-containing arm could have emerged later as the template reading head. On the protein side of the aminoacylation reaction, the class-defining catalytic domain is envisioned as the ancestral enzyme, with domains making tRNA-specific contacts subsequently added.
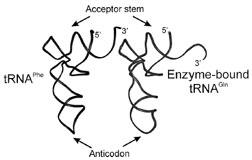 |
| Figure 3 Distortion of tRNAGln in complex with GlnRS. The cocrystal structure of GlnRS:tRNAGln revealed a distortion at the 3´-end of the tRNA (right) in contrast to uncomplexed tRNAPhe (left). This uncoupling of the first acceptor helix base pair and folding back of the CCA end of the tRNA is apparently necessary for the tRNA to reach the glutaminyl adenylate bound in the enzyme active site. |
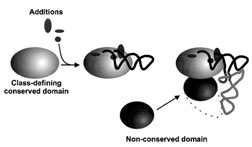 |
| Figure 4 Domain organization of synthetases and tRNAs. The class-defining domain of the aminoacy l–tRNA synthetase contains conserved structural features and contacts the acceptor arm of tRNA. These contacts are mediated by additions to the classdefining catalytic domain. Appended nonconserved protein domains interact with other portions of the tRNA, including in many cases the anticodon (as indicated by the dotted line). [From Schimmel, P., and Ribas de Pouplana, L. (1995). Cell 81, 983–986.] |
H. Novel Functions of AARSs
In addition to their critical role in protein synthesis, it has become clear that AARSs are involved in several other cellular pathways. Some AARSs regulate their own transcription and translation, while others contribute to splicing activities in mitochondria. Nuclear aminoacylation of tRNAs by imported AARSs is thought to be a quality control mechanism to ensure that only mature, fully active tRNAs are released efficiently to the cytoplasm for protein synthesis. Programmed cell death (apoptosis) also appears to have an AARS component—human tyrosyl–tRNAsynthetase can be proteolytically cleaved into two polypeptides with distinct cytokine activities, despite the lack of such activity in the full-length TyrRS. It is likely that in time many more nontranslational functions of AARS will be identified.




