Translation Initiation
Initiation of protein synthesis requires assembly of the ribosomal subunits, messenger RNA, and initiator tRNA at the start codon. This organization of translational components is facilitated by protein initiation factors (Fig. 5).A. The Message
A combination of nucleotide signals identifies the beginning of anmRNAsequence to be translated into its protein product. The nucleotide triple AUG is the start codon that directs the ribosome to begin reading an mRNA and orients the message in the right frame (for example, . . . CUA GUG CAC C. . . rather than . . . C UAG UGC ACC. . . , whichwould be a different protein). However,AUGis also the codon for insertion of the amino acid methionine into the body of the polypeptide chain. What distinguishes the start AUG from other identical codons elsewhere in the message? A stretch of 3–10 nucleotides located about 10 nucleotides upstream (in the 5´-direction) from the start codon is called the Shine–Dalgarno sequence, after the researchers who identified it. This sequence is rich in A and G nucleotides, and is partially complementary to a short region of U and C nucleotides near the 3´-end of an RNA molecule embedded within the ribosomal small subunit. Such complementarity positions the incoming message properly on the ribosome, so that the start codon is in the ribosomal decoding site for initiation of protein synthesis. Once translation begins, the rest of the codons in the message need to be read, so the base-pairing interaction between mRNA and rRNA at the Shine–Dalgarno sequence must be transient.
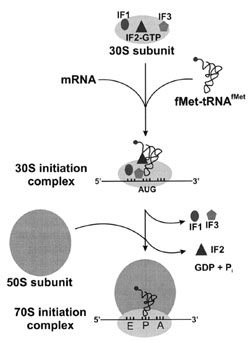 |
| Figure 5 Translation initiation. In prokaryotes, three initiation factors are responsible for assembling the initiation complex prior to decoding of a message. The mRNA and initiator tRNA bind to the small ribosomal subunit in random order, with IF2 selectively binding initiator tRNA. Hydrolysis of IF2-bound GTP promotes formation of the 30S initiation complex. Initiation Factors 1 and 3 leave the complex, and the large ribosomal subunit binds to form the 70S initiation complex with release of IF2. |
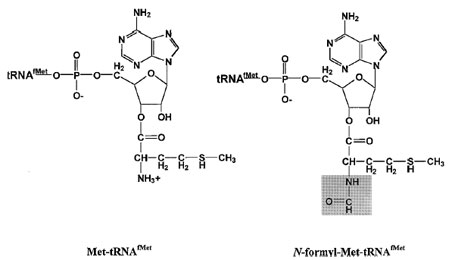 |
| Figure 6 N-formyl methionyl–tRNAfMet. Sequence and structural features target methionylated tRNAfMet for formylation at the free amino group. This modification of the aminoacylated tRNA blocks the amino group and introduces an amide bond (highlighted in gray). Together with the uniquely rigid anticodon stem, these elements allow recognition by IF2 and placement of the initiator molecule in the ribosomal P-site for interaction with the AUG start codon of mRNA. |
B. Initiator tRNA
The tRNA that recognizes the AUG start codon and the amino acid attached to it are also distinct from the Met– tRNAMet base-paired to internal AUG codons. This initiator tRNA contains the CAU anticodon necessary for recognition of the start codon, but several sequence and structural differences distinguish it from the elongator tRNA that inserts methionine into internal positions of the polypeptide. In bacteria, both elongator and initiator tRNAs areaminoacylatedbymethionyl–tRNAsynthetase, but the methionylated initiator tRNA undergoes further processing prior to its transport to the ribosome. The enzyme fMet–tRNA transformylase modifies the amino group of the tRNA-attached methionine residue, using N10-formyltetrahydrofolate as a formyl donor. One feature that the transformylase enzyme recognizes is a mismatched base pair at the first position of the initiator tRNA (tRNAfMet) acceptor stem. The elongator tRNA (tRNAMet) molecule has a canonical G:C base pair at the first position of the acceptor stem, while tRNAfMet contains a C:A pair. Base substitutions that produce a strong base pair at this position in tRNAfMet significantly decrease formylation of Met–tRNAfMet.
Modification of the aminoacylated initiator tRNA to produce formyl–methionine–tRNAfMet blocks the amino group of methionine and introduces an amide bond (Fig. 6). The lack of a free amino group in fMet–tRNAfMet prevents its insertion into a protein anywhere but at the N-terminal position. Furthermore, the presence of an amide bond targets fMet–tRNAfMet for the P-site of the ribosome. Only the initiator tRNA enters the Psite directly—all other aminoacyl–tRNAs enter the ribosome at the A-site and are moved to the P-site after peptide bond formation. This targeting is achieved both by the presence of the amide bond of fMet–tRNAfMet and by the uniquely rigid anticodon stem of the initiator tRNA, which contains three G:C base pairs. Progressive substitution of these three G:C pairs has been shown to weaken binding of the initiator tRNA to the P-site.
C. Initiation Factors
Assembly of the ribosomal subunits, mRNA, and initiator tRNA into a complex ready for protein synthesis requires several proteins called initiation factors. In prokaryotes, three initiation factors (IFs) transiently associate with the components of the translational machinery: IF1, IF2, and IF3. (In eukaryotes, more factors are required but the overall initiation process is similar with a few exceptions described below.) Table II summarizes the properties of E. coli initiation factors as well as protein factors involved in elongation and termination.
| Protein factor | Size in E. coli (kD) | Function | |
|---|---|---|---|
| Initiation | IF1 | 9 | Stimulates IF2/IF3 binding and functions |
| IF2 | 97 | GTPase, binds fMet–tRNAfMet | |
| IF3 | 22 | Prevents subunit association | |
| Elongation | EF-Tu | 43 | GTPase, transports AA–tRNA to ribosome |
| EF-G | 74 | GTPase, stimulates translocation of tRNA on ribosome | |
| EF-Ts | 77 | Nucleotide exchange factor (EF-Tu: GDP to EF-Tu:GTP) | |
| Termination | RF1 | 36 | Promotes polypeptide release at stop codons UAG and UAA |
| RF2 | 38 | Promotes polypeptide release at stop codons UGA and UAA | |
| RF3 | 46 | GTPase, stimulates RF1 and RF2 function | |
| RRF | 20 | Promotes dissociation of post-termination complex |
Biochemical studies determined that each initiation factor has a distinct high-affinity binding site on the 30S subunit. Based on in vivo concentrations and affinities, the IFs most probably do not exist free in solution but are predominantly bound to 30S subunits. When cellular concentrations of mRNA and fMet–tRNAfMet are high enough, these RNAs bind the small subunit (containing a single molecule of each IF) in random order. In this ternary complex, the mRNA and tRNA are not in contact with each other, but a kinetic rearrangement triggered by the IFs positions the initiator tRNA in the part of the P-sitecontributedbythe30Sparticleandpromotes thefirst codon–anticodon interaction. This 30S initiation complex can either dissociate into its individual components or bind the 50S subunit (with IF2-mediated hydrolysis of GTP) to form a 70S initiation complex.
Each factor plays a particular role in translation initiation, acting together to form 30S initiation complexes that can proceed toward elongation. The precise function of IF1 is not known, although it does stimulate the binding and activities of IF2 and IF3. It binds to 30S subunits directly, and in combination with the other factors promotes the formation of 30S initiation complexes. Once the 30S complex is formed, IF1 is ejected along with IF3. To date there is no conclusive evidence that the IFs affect the Shine–Dalgarno interaction between the mRNA and rRNA.
The most active role in initiation seems to belong to IF2, which has binding sites for fMet–tRNAfMet, GTP, and both ribosomal subunits. Initiation Factor 2 promotes the association of fMet–tRNAfMet with the small subunit and, in particular, recognizes the blocked amino group of this tRNA. The activities of IF2 are partitioned between two domains of the protein—the C-terminal domain is thought to be responsible for initiator tRNA binding, while the central domaincontainsaGTPasefunction. Whichportion of IF2 binds the 30S subunit remains to be determined. Hydrolysis of GTP to GDP occurs only upon 50S binding to the ternary complex; IF2 has no GTPase activity in the absence of the ribosome. As GTP hydrolysis is the final step in initiation, a conformational change in the ribosome is thought to eliminate IF2 from the initiation complex, further orient the initiator tRNA in the P-site, or otherwise contribute to a kinetic proofreading mechanism.
Initiation Factors 2 and 3 have seemingly opposing functions. While IF2 promotes the binding of tRNA to the 30S subunit, IF3 can be considered the subunit “antiassociation” factor because it increases the rates of subunit exchange and complex dissociation. In fact the two functions cooperate to facilitate formation of the correct 30S initiation complex—IF2 preferentially enhances the binding of the amino-blocked initiator tRNA and IF3 specifically increases the rate of noninitiator tRNA dissociation from the ternary complex. Initiation Factor 3 also contributes to the fidelity of translation by confirming the codon–anticodon interaction on the 30S subunit.
D. Eukaryotic Initiation
Many more protein factors are involved in eukaryotic initiation; some systems contain more than 10 initiation factors. Particular features of translation initiation are also different in the higher organisms. Most notably, prokaryotic ribosomes can initiate internally on an mRNA (even on circular RNAs), while in eukaryotes a “preinitiation” complex binds to the 5´-end of the mRNA and then progresses to an initiation complex. Eukaryotic mRNAs are capped at their 5´-end with a 7-methylguanosine triphosphate structure, and one of the eukaryotic initiation factors binds this capped end. The preinitiation complex then moves along themRNAand initiates translation at the first AUG codon it comes to. Consistent with this scanning mechanism is the observation that eukaryotic mRNAs do not contain Shine–Dalgarno-like sequences.




