Elongation
The heart of protein biosynthesis lies in the elongation cycle, with its sequential decoding of mRNA codons to assemble the useful portion of the polypeptide. Elongation can be further broken down into three phases— aminoacyl–tRNA decoding, peptide bond formation, and translocation of the new peptidyl–tRNA (Fig. 7).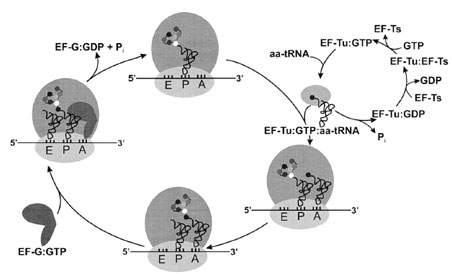 |
| Figure 7 The elongation cycle. Aminoacylated tRNAs are transported to the ribosome by EF-Tu, and they are positioned in the ribosomal A-site upon hydrolysis of EF-Tu-bound GTP. Nucleotide exchange is catalyzed by EF-Ts. Following peptide bond formation, the GTPase EF-G triggers translocation of the peptidyl–tRNA from the A-site to the P-site; the empty (deacylated) tRNA exits the ribosome by way of the E-site. The mRNA moves the length of one codon in the 3´ direction, probably pulled through the ribosome by tRNA translocation. |
A. Decoding According to Base Pairing
Comparison of the anticodon of the incoming AA–tRNA with the corresponding mRNA codon takes place at the decoding center of the ribosome, located on the small subunit. Peptide bond formation occurs on the large ribosomal subunit, at the peptidyl transferase center. Thus, this segregation of functions parallels the two arms of the tRNA. The anticodon portion of the tRNA binds to the small subunit, where the genetic message is read. The acceptor arm of tRNA (with its attached amino acid) contacts the large subunit, where catalysis occurs.
Although the synthesis of a peptide bond is the key step in translation, this is the easiest part of protein synthesis. Once the amino group of an aminoacyl–tRNA is properly positioned close enough to the carbonyl group of a peptidyl–tRNA, peptide bond formation through nucleophilic attack is energetically favorable. The ribosome can be considered as a single enzyme whose function is to catalyze peptide bond formation.
B. Elongation Factors
Addition of each incoming amino acid requires the cooperation of three elongation factors. Elongation factor Tu (EF-Tu) is the most abundant protein in E. coli, with about 100,000 copies per cell, or 5% of the cell’s protein. This protein is a GTPase, and the EF-Tu:GTP complex specifically binds aminoacyl–tRNAs (AA–tRNAs). Formation of the ternary complex (EF-Tu:GTP:AA–tRNA) protects the ester bond (linking the amino acid to its cognate tRNA) from hydrolysis, and transports the AA–tRNA to the ribosomal A-site. Once the correct codon–anticodon interaction is confirmed, ribosome-triggered hydrolysis of EF-Tu-bound GTP occurs. EF-Tu:GDP is then released from the ribosome, and the AA–tRNA occupies the Asite. While EF-Tu transports all elongator tRNAs aminoacylated with natural amino acids to the ribosome, this factor has negligible affinity for formylated or nonformylated tRNAfMet. The unpaired first position in the tRNAfMet acceptor stem helix apparently is a negative recognition element for EF-Tu:GTP, because this element prevents the initiator tRNA from pairing with internal AUG or GUG codons.
The elongation factor EF-Ts is a nucleotide exchange factor that regenerates active EF-Tu:GTP (from EFTu: GDP) for binding subsequent AA–tRNAs following GTP hydrolysis. Before their functions were known, elongation factors Tu and Ts were named for their observed thermal stabilities in vitro—Tu indicates that this protein is Temperature unstable, while Ts stands for Temperature stable. In eukaryotes, the two subunits of elongation factor EF-1 perform the functions of EF-Tu and EF-Ts.
Once the A-site is occupied by the incomingAA–tRNA, nucleophilic attack on the peptidyl–tRNA by the AA– tRNA occurs. The condensation reaction produces a new peptide bond and lengthens the polypeptide chain by one amino acid (Fig. 8). As a result, the growing protein is now attached to the A-site tRNA; this addition to and transfer of the polypeptide chain is called transpeptidation.
Following formation of the peptide bond, a major rearrangement of components in the functional center of the ribosome must take place. Because the most recently entered tRNA has become the peptidyl–tRNA, it must be moved from the A-site to the P-site. The former peptidyl–tRNA has been deacylated and needs to vacate the P-site. Finally the mRNA must move three nucleotides further in the 3´-direction so that the next codon can be read. The concerted movement of tRNAs and mRNA at the end of each elongation round is called translocation, and is catalyzed by elongation factor G (EF-G), another of the GTPase proteins in the translational machinery.
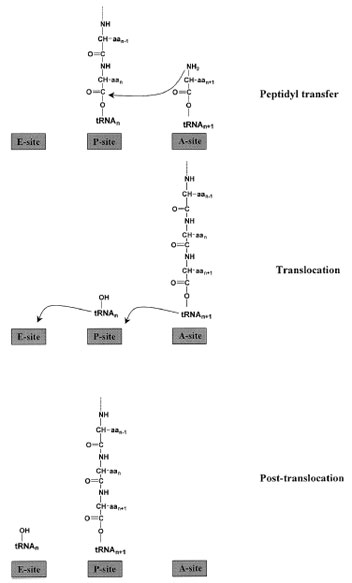 |
| Figure 8 Steps in elongation. With the peptidyl–tRNA bound in the P-site and the incoming aminoacyl–tRNA in the A-site, the peptidyl transferase activity of the large ribosomal subunit catalyzes peptide bond formation. The growing polypeptide chain is then attached to the A-site-tRNA, and the deacylated tRNA is in the P-site. Elongation factor G facilitates translocation of the peptidyl–tRNA to the P-site and the empty tRNA to the E-site prior to its release from the ribosome. |
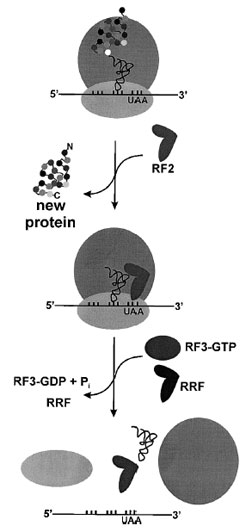 |
| Figure 9 Termination of protein synthesis and ribosome recycling. In prokaryotes, RF1 hydrolyzes the newly synthesized protein at stop codons UAG and UAA, while RF2 recognizes stop codons UGA and UAA. The GTPase RF3 stimulates release of either RF1 or RF2. In eukaryotes a single protein recognizes all stop codons. The final step of translation is dissociation of the inactive 70S complex, stimulated by the ribosome recycling factor (RRF). |




