Transfer RNAs
Although an mRNA nucleotide sequence dictates the polypeptide sequence to be made, mRNAs do not directly recognize amino acids. Amino acids are instead linked to transfer RNA (tRNA) “adaptor” molecules, which serve as reading heads to decipher the codons of mRNA through base-pairing complementarity (Fig. 2).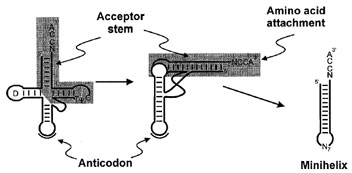 |
| Figure 2 Transfer RNA folding. The tRNA cloverleaf secondary structure representation (left) folds into an L-shaped structure consisting of two domains (center). The highly conserved D and TΨC loops are indicated. Tertiary interactions, including unusual base pairs and base triples facilitate and stabilize formation of the corner of the L. The anticodon is separated approximately 75 Å from the site of amino acid attachment. The RNA minihelix domain is highlighted by shading, while the second domain containing the anticodon and D-arm is unshaded. Minihelix RNAs are in many systems substrates for aminoacylation by aminoacyl-tRNA synthetases. |
A. Conserved Features of tRNAs
Transfer RNAs typically contain approximately 76 nucleotides and have a molecular mass of about 25 kD. The characteristic “cloverleaf ” secondary structure representation of tRNA was predicted based on regions of base complementarity, and the determination of hundreds of tRNA sequences demonstrated that such a folding pattern is conserved. Several sequence and structural features are alsoconsistentlypresent intRNAs fromall organisms. The sequence at the 3´-end of tRNAs is always –CCA, with a free hydroxyl group on the terminal adenosine that is the siteof aminoacidattachment. Nucleotides near thetermini hybridize to make the 7-base-pair (bp) acceptor stem.
The other arms of the tRNA cloverleaf also have distinctive conserved features. The modified base dihydrouridine (D) is typically present in the loop that closes off a short 3- or 4-bp stem following the acceptor stem. This stem and loop are therefore called the D-arm. The anticodon arm consists of a 5-bp helix closed by a loop that contains the trinucleotide anticodon. Following the anticodon arm is the variable loop, which can contain 3–21 nucleotides, with a stem as long as 7 bp, depending on the particular tRNA. The modified bases pseudouridine (Ψ) and ribothymidine (T) are usually present in the loop of the TΨC arm, so named because of the presence of this highly conserved sequence.
B. The L-Shaped Structure of tRNAs
In three dimensions, tRNAs fold into an L-shaped structure in which the acceptor stem and TΨC arm coaxially stack to form one part of the L known as the minihelix, and the D and anticodon arms likewise stack to form the other part of the molecule. This structure is facilitated and stabilized by tertiary interactions at the corner of the L that bring together the D and variable loops. The nucleotides involved in these interactions are typically invariant or semi-invariant, indicating that the tRNA L shape is universal. While most base pairs in tRNA helices are canonical Watson–Crick pairs, the tertiary interactions at the corner of the L make use of some unusual hydrogen-bonding conformations. For example, nearly all tRNAs contain a U8:A14 reverse Hoogsteen base pair, and several base triples (where three bases are paired together) are also typically present at the core of the structure.
The two portions of the L can be considered distinct domains with separate contributions to protein synthesis. The minihelix containing the acceptor arm includes the site of amino acid attachment (the 3´-OH). It is considered by many investigators to be related to the historical or early form of tRNA. The anticodon trinucleotide is located at the other end of the L, approximately 75 Å away. This separation between anticodon and amino acid on the tRNA is paralleled in the mRNA decoding and peptide bond formation events that occur on two different subunits of the ribosome. Furthermore, the domains of tRNA can be physically separated such that an isolated acceptor stem (“minihelix”) can in many cases accept its specified amino acid, while an anticodon stem-loop helix can bind to the ribosome-bound mRNA.
C. Codon–Anticodon Interactions
Translation of a genetic message into its protein product depends on base pairing interactions between mRNA codon and tRNA anticodon. The 64 trinucleotide codons are more than sufficient to fully determine all 20 amino acids as well as one start and three stop codons. Thus, the code is degenerate, with many amino acids having more than one codon. This codon degeneracy is primarily due to variation in the third position of the trinucleotide, as shown in Table I. Although many organisms have more than one tRNA molecule per amino acid (these are “isoacceptors”), in many cases a specific tRNA recognizes more than one codon. Non-Watson–Crick base pairs are permitted at the third position, because of room for some structural flexibility or “wobble” in the pairing geometry. For example, a U in the third position (5´ nucleotide) of the anticodon can base pair with anA(Watson–Crick pair) or a G (wobble pair) in the codon. Several tRNAs contain an inosine (I) nucleotide at the third anticodon position; inosine forms a standard base pair with C and wobble pairs with U and A.




