Gene Regulation in Eukaryotes
Gene Regulation in Eukaryotes
There are a number of different phenomena in eukaryotic cells that can serve as control points, and the following are a few examples.
Transcriptional Control This may
be the most important mechanism. Transcription factors are molecules
that may have a positive or a negative
effect on transcription of RNA from the
DNA of the target genes. The factors
may act within the cells that produce
them or they may be transported to
different parts of the body prior to
action. An example of a positive transcription
factor is a steroid receptor.
Steroid hormones produced by endocrine
glands elsewhere in the body
enter the cell and bind with a receptor
protein in the nucleus. The steroidreceptor
complex then binds with
DNA near the target gene.
Progesterone, for example, binds with
a nuclear receptor in cells of the
chicken oviduct; the hormone-receptor
complex then activates the transcription
of genes encoding egg albumin
and other substances.
Translational Control Genes can be transcribed and the mRNA sequestered in some way so that translation is delayed. This commonly happens in the development of eggs of many animals. The oocyte accumulates large quantities of messenger RNA during its development, then fertilization activates metabolism and initiates translation of maternal mRNA.
Gene Rearrangement Vertebrates contain cells called lymphocytes that bear genes coding for proteins called antibodies. Each type of antibody has the capacity to bind specifically with a particular foreign substance (antigen). Because the number of different antigens is enormous, the genetic diversity of antibody genes must be equally great. One source of this diversity is rearrangement of DNA sequences coding for the antibodies during the development of lymphocytes.
DNA Modification An important mechanism for turning genes off appears to be methylation of cytosine residues, that is, adding a methyl group (CH3−) to the carbon in the 5 position in the cytosine ring (Figure 5-22A). This usually happens when the cytosine is next to a guanine residue; thus, the bases in the complementary DNA strand would also be a cytosine and a guanine (Figure 5-22B). When the DNA is replicated, an enzyme recognizes the CG sequence and quickly methylates the daughter strand, maintaining the gene in an inactive state.
There are a number of different phenomena in eukaryotic cells that can serve as control points, and the following are a few examples.
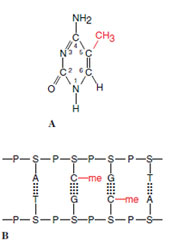 |
| Figure 5-22 Some genes in eukaryotes are turned off by the methylation of some cytosine residues in the chain. A, Structure of 5- methyl cytosine. B, Cytosine residues next to guanine are those that are methylated in a strand, thus allowing both strands to be symmetrically methylated. |
Translational Control Genes can be transcribed and the mRNA sequestered in some way so that translation is delayed. This commonly happens in the development of eggs of many animals. The oocyte accumulates large quantities of messenger RNA during its development, then fertilization activates metabolism and initiates translation of maternal mRNA.
Gene Rearrangement Vertebrates contain cells called lymphocytes that bear genes coding for proteins called antibodies. Each type of antibody has the capacity to bind specifically with a particular foreign substance (antigen). Because the number of different antigens is enormous, the genetic diversity of antibody genes must be equally great. One source of this diversity is rearrangement of DNA sequences coding for the antibodies during the development of lymphocytes.
DNA Modification An important mechanism for turning genes off appears to be methylation of cytosine residues, that is, adding a methyl group (CH3−) to the carbon in the 5 position in the cytosine ring (Figure 5-22A). This usually happens when the cytosine is next to a guanine residue; thus, the bases in the complementary DNA strand would also be a cytosine and a guanine (Figure 5-22B). When the DNA is replicated, an enzyme recognizes the CG sequence and quickly methylates the daughter strand, maintaining the gene in an inactive state.




