Eukaryotic DNA polymerases
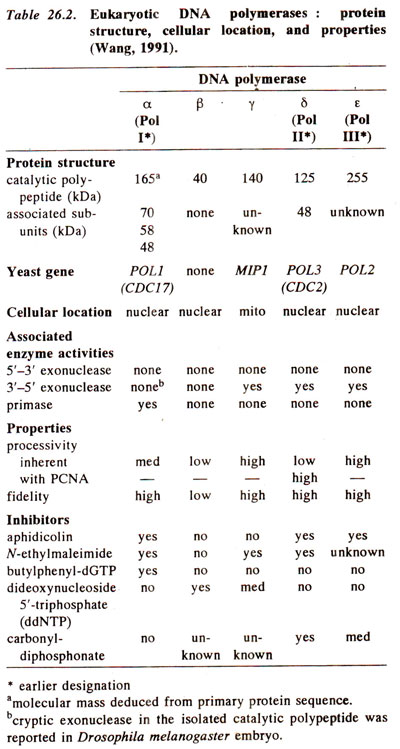
Five different eukaryotic DNA polymerases have been identified, from materials like yeast cells, rat liver, human cell cultures and tumour cells. These include the following : (i) DNA potymerase a, a relatively high molecular weight enzyme, also called cytoplasmic polymerase (also appropriately called large polymerase, since it is also found in the nucleus and since another small nuclear polymerase is also available in cytoplasm); (ii) DNA polymerase β or nuclear polymerase (also called small polymerase), found only in vertebrates, and not in budding yeast nor in other lower eukaryotes; (iii) DNA polymerase γ or mitochondrial polymerase, encoded in the nucleus; (iv) DNA polymerase δ, another novel enzyme reported from mammalian cells, is PCNA dependent for DNA-synthetic processivity (PCNA = proliferating cell nuclear antigen; processivity = property of continuous synthesis); (v) DNA polymerase ε (earlier described as DNA polymerase δ11), also purified and reported from mammalian Hela cells and budding yeast, is PCNA independent. In some other cases, a distinction between DNA polymerase δ and DNA polymerase ε has not been possible. Another calf thymus enzyme designated as DNA polymerase 61 represented DNA polymerase αwith 3'-5' exonuclease activity. The large DNA polymerase a is the predominant DNA polymerase in eukaryotic cells and was believed for long time to be the only enzyme involved in DNA replication.
However, both DNA polymerase αand DNA polymerase δ are now known to take part in eukaryotic DNA replication (see later for details). DNA polymerase αhas four subunits and an appropriate designation for this enzyme is DNA polymerase α/primase. Three other enzymes which are found associated with large DNA polymerase αinclude thymidine kinase, ribonucleotide reductase and thymidylate synthetase. They are also believed to be involved in DNA replication.
Nuclear small DNA polymerase βresembles large DNA polymerase αin its replicative activity, but differs in some other respects. Its quantity remains unchanged, while that of large polymerase changes with need. It is speculated that small enzyme may be involved in repair and recombination. The differences between these polymerases are shown in Table 26.2.
Nuclear small DNA polymerase βresembles large DNA polymerase αin its replicative activity, but differs in some other respects. Its quantity remains unchanged, while that of large polymerase changes with need. It is speculated that small enzyme may be involved in repair and recombination. The differences between these polymerases are shown in Table 26.2.