Isoquinoline Alkaloid Biosynthesis
Isoquinoline alkaloids are a large and diverse group of alkaloids with~2500 defined
structures. They include the analgesic morphine from
Papaver somniferum L.,
the antigout colchicine from
Colchicum autumnale L., the emetic and antiamoebic
emetine from
Cephaelis ipecacuanha (Brot.) A. Rich., the skeletal muscle relaxant
tubocurarine from
Strychonos toxifera Bentham, and the antimicrobial compounds
berberine and sanguinarine from divergent plant species including
Berberis spp.
and
Sanguinaria spp., many of which are used as pharmaceuticals.
Isoquinoline alkaloid biosynthesis begins with the conversion of tyrosine to
both dopamine and 4-hydroxyphenylacetaldehyde by decarboxylation, orthohydroxylation,
and deamination (Fig. 11.2; Facchini, 2001). Among these early
steps, only tyrosine/dopa decarboxylase (TYDC; an aromatic L-amino acid decarboxylase),
which converts tyrosine and dopa to their corresponding amines, has
been purified and characterized. This small family of genes (~15 genes) was
isolated from opium poppy (
P. somniferum) and each subfamily has been shown
to have distinct developmental and inducible expression patterns (Facchini and
De Luca, 1994, 1995; Park
et al., 1999). Members of the TYDC gene family are
classified into two groups (TYDC1 and TYDC2) that are differentially expressed in
opium poppy. In the mature plant, TYDC2-like transcripts are predominant in the
stem and are also present in roots, whereas TYDC1-like transcripts are abundant
only in roots. The localization of TYDC transcripts in the phloem is consistent with
the expected developmental origin of laticifers, which are specialized internal
secretory cells that accompany vascular tissues in all organs of select species
and contain alkaloid-rich latex in aerial organs (Facchini and De Luca, 1995).
Dopamine and 4-hydroxyphenylacetaldehyde are condensed by norcoclaurine
synthase (NCS) to yield (S)-norcoclaurine, which is the central precursor to all
isoquinoline alkaloids. Recently, NCS has been purified and characterized
(Samanani and Facchini, 2002) from cultured
Thalictrum flavum spp., and a
TfNCS cDNA belonging to PR10 family was isolated from
T. flavum (Samanani
et al., 2004), whereas a novel dioxygenase-like protein (CjNCS) from cultured
Coptis japonica cells was also shown to catalyze this NCS reaction (Minami
et al., 2007). The presence of TYDC (Facchini, 2001) and TfNCS (Samanani
et al., 2004)
and CjNCS homologues (Minami
et al., 2007) in
Arabidopsis or rice suggests
that these genes either have other basic biological roles or that the isoquinoline
biosynthesis pathway is relatively universal in the plant kingdom, although
sequence homology of TfNCS or CjNCS with
Arabidopsis or rice homologues
were relatively low (less than 20% in amino acid basis) (Liscombe
et al., 2005;
Minami
et al., 2007) and no isoquinoline alkaloid has been found in
Arabidopsis or
rice. Strictosidine synthase (STR) (the key reaction in terpenoid indole alkaloid)-
like genes has also been found in animals and
Arabidopsis (De Luca and Laflamme,
2001).
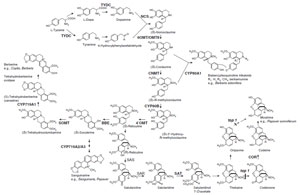 |
| FIGURE 11.2 Biosynthetic pathways to various
isoquinoline alkaloids. Unbroken arrows indicate
single enzymatic conversions and broken arrows
indicate multiple enzymatic steps. Enzymes for
which the corresponding genes have been cloned
are indicated in bold. TYDC, tyrosine/dopa
decarboxylase; NCS, norcoclaurine synthase;
6OMT, norcoclaurine 6-O-methyltransferase;
CNMT, coclaurine N-methyltransferase;
CYP80B1,
N-methylcoclaurine 3'-hydroxylase;
4'OMT, 3' hydroxy N-methylcoclaurine
4'-O-methyltransferase; BBE, berberine bridge
enzyme; CYP719A1,
canadine synthase
(methylenedioxy bridge-forming enzyme);
SOMT, scoulerine 9-O-methyltransferase; SAS,
salutaridine synthase; SAR, salutaridine
reductase; SAT, acetylcoenzyme
A:salutaridinol-7-O-acetyltransferase; COR,
codeinone reductase; CYP80A1, berbamunine
synthase. |
(S)-Norcoclaurine is sequentially converted to coclaurine by S-adenosyl methionine
(SAM)-dependent norcoclaurine 6-O-methyltransferase (6OMT) (Morishige
et al., 2000), to N-methylcoclaurine by coclaurine N-methyltransferase (Choi
et al., 2002), to 3'-hydroxy-N-methyl coclaurine by P450 hydroxylase (Pauli and
Kutchan, 1998), and then to (S)-reticuline by 3'-hydroxy N-methylcoclaurine
4'-O-methyltransferase (4'OMT; see Fig. 11.2; Morishige
et al., 2000). All of the
cDNAs for these reactions have been isolated and functional recombinant
proteins subsequently produced. Detailed biochemical studies using recombinant
enzymes have shown their strict reaction specificities, and these enzymes
regulate biosynthesis sequentially and in a coordinated manner. For example,
CNMT prefers coclaurine than 6-O-methylnorlaudanosoline and 4'OMT prefers
an N-methylated substrate, which suggest that the pathway in Fig. 11.2 is
preferable to a sequence of N-methylation, hydroxylation, and 4'-O-methylation.
On the other hand,
Thalictrum cells may show some variation since
Thalictrum O-methyltransferases can form heterodimers and exhibit broad substrate specificity
(Frick and Kutchan, 1999). Current data also indicate that all of these enzymes,
except the membrane-bound P450 CYP80B1, are located in the cytosol.
While dimeric bisbenzylisoquinoline alkaloids, such as berbamunine and
tubocurarine, are produced from the intermediates of the (S)-reticuline pathway
by the action of a phenol coupling P450-dependent oxidase (berbamunine synthase,
CYP80A1) (Kraus andKutchan, 1995), reticuline is the central intermediate in branch
pathways that lead to benzophenanthridine alkaloids (e.g., sanguinarine and marcarpine),
protoberberine alkaloids (e.g., berberine and palmatine), and morphinan
alkaloids (e.g., morphine and codeine) (Fig. 11.2). Many of the enzymes involved in
these branch pathways have been purified and the corresponding cDNAs have
been cloned.
The first committed step in the biosynthesis of benzophenanthridine, a protoberberine
alkaloid, involves conversion of the N-methyl group of (S)-reticuline into
the methylene bridge moiety of (S)-scoulerine by the berberine bridge enzyme (BBE) (Dittrich and Kutchan, 1991). This unique enzyme is soluble but localized
in vesicles (Bock
et al., 2002). Immunocytological staining of
P. somniferum tissue
with antibodies against BBE led to a characteristic labeling of electron-dense
aggregates in idioblasts that are not connected to the laticifer system, which
demonstrates that benzophenanthridine and morphine biosyntheses show strict
cytological separation within this plant (Bock
et al., 2002).
In benzophenanthridine alkaloid biosynthesis, (S)-scoulerine can be converted
to (S)-stylopine by two P450-dependent oxidases, (S)-cheilanthifoline and
(S)-stylopine synthase, which result in the formation of two methylenedioxy groups
(not shown) (see Facchini, 2001; Ikezawa
et al., 2007). On the other hand, in protoberberine
biosynthesis, (S)-scoulerine is converted to (S) tetrahydrocolumbamine by
the SAM-dependent scoulerine 9-O-methyltransferase (SOMT) (Takeshita
et al., 1995) and then to tetrahydroberberine (canadine) by a P450-dependent canadine
synthase (CDS or CYP719A1) (Ikezawa
et al., 2003). The isolation and characterization
of these enzymes and the corresponding cDNAs have confirmed that berberine
biosynthesis proceeds via canadine and not via columbamine. Again, the enzyme
substrate specificity shows a clear preference for this pathway.While the hydrophobic
N-terminal region of SOMT suggests that this enzyme may be targeted to the
membrane fraction, its localization in both the cytosol (Muemmler
et al., 1985) and
within the lumen of alkaloid-specific vesicles (Galneder
et al., 1988) has been
reported. Note that the CYP719A1 family as well as CYP80 were not found in
Arabidopsis and rice and that these members of the cytochrome P450 superfamily
are unique for benzylisoquinoline alkaloid biosynthesis (Nelson
et al., 2004).
In morphinan alkaloid biosynthesis, (S)-reticuline is converted to its (R)-enantiomer
via the stereospecific reduction of 1,2-dehydroreticuline with NADPHdependent
cytosolic 1,2-dehydroreticuline reductase. Subsequent intramolecular
carbon–carbon phenol coupling of (R)-reticuline by a P450-dependent salutaridine
synthase results in the formation of salutaridine. Salutaridine: NADPH
7-oxidoreductase then reduces salutaridine to (7S)-salutaridinol. Transformation
of salutaridinol into thebaine involves the closure of an oxide bridge between C-4
and C-5 by acetylcoenzyme A:salutaridinol-7-O-acetyltransferase (SAT) (Grothe
et al., 2001). Furthermore, thebaine can be converted to codeinone and then
reduced to codeine by cytosolic NADPH-dependent codeinone reductase (COR)
(Unterlinner
et al., 1999). Finally, codeine is demethylated to give morphine.
Interestingly, COR genes have been found in some
Papaver spp. that do not
produce morphine (Unterlinner
et al., 1999), whereas SAT transcript was detected
in
Papaver spp. that accumulate alkaloids with a morphinan nucleus, consistent
with the expected distribution (Grothe
et al., 2001; Unterlinner
et al., 1999). The
recent isolation of the top1 mutant from poppy, and the demonstration of the
activity of the protein, has illustrated that thebaine can be demethylated in two
steps either through codeinone or oripavine to morphine (Millgate
et al., 2004).
Northern blot analysis using the eight available genes in morphinan alkaloid
biosynthesis showed that while all of the transcripts are detected in every organ,
the highest levels are seen in stems and flower buds and the lowest levels are seen
in leaves (Millgate
et al., 2004; Unterlinner
et al., 1999). The accumulation of each
transcript, with the exception of COR, was markedly induced in response to treatment with an elicitor or wounding of cultured cells. All known enzymes in
the morphine pathway have been detected in cultured cells derived from the fruit
capsule (Facchini and Park, 2003; Grothe
et al., 2001; Huang and Kutchan, 2000;
Unterlinner
et al., 1999). Conversely, different cell type-specific localizations of
biosynthetic enzymes have been reported in the capsule and stem of intact
opium poppy plants.
In situ localization of alkaloid biosynthetic gene transcripts
indicated seven biosynthetic enzymes: 6OMT, CNMT, CYP80B, 4'OMT and BBE
involved in reticuline biosynthesis, and SAT and COR in morphine pathway.
These proteins have apparently been localized to sieve elements in opium
poppy and the corresponding gene transcripts to adjacent phloem companion
cells (Bird
et al., 2003; Facchini and St-Pierre, 2005). In contrast, a different immunocytochemical
analysis clearly showed that 4'OMT and SAT were located in
phloem parenchyma cells in vascular bundles. COR, catalyzing the penultimate
step in morphine biosynthesis, was localized to laticifers, the site of morphinan
alkaloid accumulation (Weid
et al., 2004). Although this discrepancy in the cell
type-specific localization of enzymes remains to be clarified, it is noteworthy that
both studies showed different localizations of the biosynthetic gene transcripts
and their corresponding enzymes.
Cell type-specific expression has recently been reported in protoberberine alkaloid
biosynthesis in
T. flavum subsp. (Samanani
et al., 2005). While gene transcripts
for biosynthetic enzyme were most abundant in rhizomes, they were also detected
at lower levels in roots and other organs. Further
In situ RNA hybridization analysis
revealed that all transcripts were mainly localized to immature endodermis
cells, the pericycle of roots, and restricted to the protoderm of leaf primordia in
rhizomes. These data and analysis of alkaloid accumulation clearly indicated that
distinct and different cell types are involved in the biosynthesis and accumulation of
benzylisoquinoline alkaloids in
T. flavum and
P. somniferum.





