Control of Microorganisms by using Antimicrobial Chemotherapy
Antimicrobial Chemotherapeutic Agents
Antimicrobial chemotherapy is the use of chemicals to inhibit or kill microorganisms in or on the host. Chemotherapy is based on selective toxicity. This means that the agent used must inhibit or kill the microorganism in question without seriously harming the host.
In order to be selectively toxic, a chemotherapeutic agent must interact with some microbial function or microbial structure that is either not present or is substantially different from that of the host. For example, in treating infections caused by prokaryotic bacteria, the agent may inhibit peptidoglycan synthesis or alter bacterial (prokaryotic) ribosomes. Human cells do not contain peptidoglycan and possess eukaryotic ribosomes. Therefore, the drug shows little if any effect on the host (selective toxicity). Eukaryotic microorganisms, on the other hand, have structures and functions more closely related to those of the host. As a result, the variety of agents selectively effective against eukaryotic microorganisms, such as fungi and protozoans, is small when compared to the number available against prokaryotes. Also keep in mind that viruses are not cells and, therefore, lack the structures and functions altered by antibiotics, so antibiotics are not effective against viruses.
Based on their origin, there are 2 general classes of antimicrobial chemotherapeutic agents:
- Antibiotics: substances produced as metabolic products of one microorganism, which inhibit or kill other microorganisms.
- Antimicrobial chemotherapeutic chemicals: chemicals synthesized in the laboratory, which can be used therapeutically on microorganisms.
Most of the major groups of antibiotics were discovered prior to 1955, and most antibiotic advances since then have come about by modifying the older forms. In fact, only 3 major groups of microorganisms have yielded useful antibiotics: the actinomycetes (filamentous, branching soil bacteria such as Streptomyces), bacteria of the genus Bacillus, and the saprophytic molds Penicillium and Cephalosporium.
To produce antibiotics, manufacturers inoculate large quantities of medium with carefully selected strains of the appropriate species of antibiotic-producing microorganism. After incubation, the drug is extracted from the medium and purified. Its activity is standardized and it is put into a form suitable for administration.
Some antimicrobial agents are cidal in action: they kill microorganisms (e.g., penicillins, cephalosporins, streptomycin, neomycin). Others are static in action: they inhibit microbial growth long enough for the body’s own defenses to remove the organisms (e.g., tetracyclines, erythromycin, sulfonamides). Antimicrobial agents also vary in their spectrum. Drugs that are effective against a variety of both Gram-positive and Gram-negative bacteria are said to be “Broad Spectrum” (e.g., tetracycline, streptomycin, cephalosporins, ampicillin, sulfonamides). Those effective against just Gram-positive bacteria, just Gramnegative bacteria, or only a few species are termed “narrow spectrum” (e.g., penicillin G, erythromycin, clindamycin, gentamycin).
If a choice is available, a narrow spectrum is preferable since it will cause less destruction to the body’s normal flora. In fact, indiscriminate use of broadspectrum antibiotics can lead to superinfection by opportunistic microorganisms, such as Candida (yeast infections) and Clostridium difficile (antibiotic-associated ulcerative colitis), when the body’s normal flora are destroyed. Other dangers from indiscriminate use of antimicrobial chemotherapeutic agents include drug toxicity, allergic reactions to the drug, and selection for resistant strains of microorganisms.
Below are examples of commonly used antimicrobial chemotherapeutic agents arranged according to their mode of action:
Antimicrobial agents that inhibit peptidoglycan synthesis
Inhibition of peptidoglycan synthesis in actively dividing bacteria results in osmotic lysis. (A list of common antimicrobial chemotherapeutic agents listed by both their generic and brand names and arranged by their mode of action.)
- Penicillins (produced by the mold Penicillium): There are several classes of penicillins:
- Natural penicillins are highly effective against Gram-positive bacteria (and very few Gram-negative bacteria) but are inactivated by the bacterial enzyme penicillinase. Examples include penicillin G, F, X, K, O, and V.
- Semisynthetic penicillins are effective against Gram-positive bacteria but are not inactivated by penicillinase. Examples include methicillin, dicloxacillin, and nafcillin.
- Semisynthetic broad-spectrum penicillins are effective against a variety of Gram-positive and Gram-negative bacteria but are inactivated by penicillinase. Examples include ampicillin, carbenicillin, oxacillin, azlocillin, mezlocillin, and piperacillin.
- Semisynthetic broad-spectrum penicillins combined with beta-lactamase inhibitors such as clavulanic acid and sulbactam. Although the clavulanic acid and sulbactam have no antimicrobial action of their own, they inhibit penicillinase, thus protecting the penicillin from degradation. Examples include amoxicillin plus clavulanic acid, ticarcillin plus clavulanic acid, and ampicillin plus sulbactam.
- Cephalosporins (produced by the mold Cephalosporium): Cephalosporins are effective against a variety of Gram-positive and Gram-negative bacteria and are resistant to penicillinase (although some can be inactivated by other beta-lactamase enzymes similar to penicillinase). Four “generations” of cephalosporins have been developed over the years in an attempt to counter bacterial resistance.
- First-generation cephalosporins include cephalothin, cephapirin, and cephalexin.
- Second-generation cephalosporins include cefamandole, cefaclor, cefazolin, cefuroxime, and cefoxitin.
- Third-generation cephalosporins include cefotaxime, cefsulodin, cefetamet, cefixime, ceftriaxone, cefoperazone, ceftazidine, and moxalactam.
- Carbapenems: Carbapenems consist of a broad-spectrum beta-lactam antibiotic to inhibit peptidoglycan synthesis combined with cilastatin sodium, an agent that prevents degradation of the antibiotic in the kidneys. An example is imipenem.
- Monobactems: Monobactems are broad-spectrum beta-lactam antibiotics resistant to beta lactamase. An example is aztreonam.
- Carbacephem: A synthetic cephalosporins. An example is loracarbef.
- Vancomycin (produced by the bacterium Streptomyces): Vancomycin and teichoplanin are glycopeptides that are effective against Gram-positive bacteria.
- Bacitracin (produced by the bacterium Bacillus): Bacitracin is used topically against Gram-positive bacteria.
Antimicrobial agents that alter the cytoplasmic membrane
Alteration of the cytoplasmic membrane of microorganisms results in leakage of cellular materials. The following is a list of common antimicrobial chemotherapeutic agents listed by both their generic and brand names and arranged by their mode of action.
- Polymyxin B (produced by the bacterium Bacillus): Polymyxin B is used for severe Pseudomonas infections.
- Amphotericin B (produced by the bacterium Streptomyces): Amphotericin B is used for systemic fungal infections.
- Nystatin (produced by the bacterium Streptomyces): Nystatin is used mainly for Candida yeast infections.
- Imidazoles (produced by the bacterium Streptomyces): The imidazoles are antifungal antibiotics used for yeast infections, dermatophytic infections, and systemic fungal infections. Examples include clotrimazole, miconazole, ketoconazole, itraconazole, and fluconazole.
Antimicrobial agents that inhibit protein synthesis
The following is a list of common antimicrobial chemotherapeutic agents listed by both their generic and brand names and arranged by their mode of action. These agents prevent bacteria from synthesizing structural proteins and enzymes.
- Agents that block transcription (prevent the synthesis of mRNA off DNA).
- Rifampins (produced by the bacterium Streptomyces): Rifampins are effective against some Gram-positive and Gram-negative bacteria and Mycobacterium tuberculosis. They inhibit the enzyme RNA polymerase.
- Agents that block translation (alter bacterial ribosomes to prevent mRNA from being translated into proteins).
- Agents such as the aminoglycosides (produced by the bacterium Streptomyces) that bind irreversibly to the 30S ribosomal subunit and prevent the 50S ribosomal subunit from attaching to the translation initiation complex. Aminoglycosides also cause a misreading of the mRNA. Examples include streptomycin, kanamycin, tobramycin, and amikacin. Most are effective against Gram-positive and Gram-negative bacteria.
- Agents that bind reversibly to the 30S ribosomal subunit in such a way that anticodons of charged tRNAs cannot align properly with the codons of the mRNA. Examples include tetracycline, minocycline, and doxycycline, produced by the bacterium Streptomyces. They are effective against a variety of Gram-positive and Gram-negative bacteria.
- Agents that bind reversibly to the 50S ribosomal subunit and block peptide bond formation during protein synthesis. Examples include lincomycin and clindamycin, produced by the bacterium Streptomyces. Most are used against Gram-positive bacteria.
- Agents that bind reversibly to the 50S ribosomal subunit and block translation by inhibiting elongation of the protein by the enzyme peptidyltransferase that forms peptide bonds between the amino acids, by preventing the ribosome from translocating down the mRNA, or both. Macrolides such as erythromycin, roxithromycin, clarithromycin, and azithromycin are examples and are used against Gram-positive bacteria and some Gram-negative bacteria.
- The oxazolidinones (linezolid) bind to the 50S ribosomal subunit and appear to interfere with the initiation of translation.
- The streptogramins (a combination of quinupristin and dalfopristin) bind to different sites on the 50S ribosomal subunit and work synergistically to inhibit translocation.
Antimicrobial agents that interfere with DNA synthesis
The following is a list of common antimicrobial chemotherapeutic agents listed by both their generic and brand names and arranged by their mode of action.
- Fluoroquinolones (synthetic chemicals): The fluoroquinolones inhibit one or more of a group of enzymes called topoisomerase, enzymes needed for bacterial nucleic acid synthesis. For example, DNA gyrase (topoisomerase II) breaks and rejoins the strands of bacterial DNA to relieve the stress of the unwinding of DNA that occurs during DNA replication and transcription. Fluoroquinolones are broad spectrum, and examples include norfloxacin, ciprofloxacin, enoxacin, levofloxacin, and trovafloxacin.
- Sulfonamides and trimethoprim (synthetic chemicals): Cotrimoxazole is a combination of sulfamethoxazole and trimethoprim. Both of these drugs block enzymes in the bacteria pathway required for the synthesis of tetrahydrofolic acid, a cofactor needed for bacteria to make the nucleotide bases thymine, guanine, uracil, and adenine.
- Metronidazole is a drug that is activated by the microbial proteins flavodoxin and feredoxin, found in microaerophilc and anaerobic bacteria and certain protozoans. Once activated, the metronidazole puts nicks in the microbial DNA strands.
Microbial Resistance to Antimicrobial Chemotherapeutic Agents
A common problem in antimicrobial chemotherapy is the development of resistant strains of bacteria. Most bacteria become resistant to antimicrobial agents by one or more of the following mechanisms:
- Producing enzymes which detoxify or inactivate the antibiotic, e.g., penicillinase and other beta-lactamases.
- Altering the target site in the bacterium to reduce or block binding of the antibiotic, e.g., producing a slightly altered ribosomal subunit that still functions but to which the drug can’t bind.
- Preventing transport of the antimicrobial agent into the bacterium, e.g., producing an altered cytoplasmic membrane or outer membrane.
- Developing an alternate metabolic pathway to bypass the metabolic step being blocked by the antimicrobial agent, e.g., overcoming drugs that resemble substrates and tie up bacterial enzymes.
- Increasing the production of a certain bacterial enzyme, e.g., overcoming drugs that resemble substrates and tie up bacterial enzymes.
These changes in the bacterium that enable it to resist the antimicrobial agent occur naturally as a result of mutation or genetic recombination of the DNA in the nucleoid, or as a result of obtaining plasmids from other bacteria. Exposure to the antimicrobial agent then selects for these resistant strains of organism.
The spread of antibiotic resistance in pathogenic bacteria is due to both direct and indirect selection. Direct selection refers to the selection of antibioticresistant pathogens at the site of infection. Indirect selection is the selection of antibiotic-resistant normal floras within an individual anytime an antibiotic is given. At a later date, these resistant normal floras may transfer resistance genes to pathogens that enter the body. In addition, these resistant normal flora may be transmitted from person to person through such means as the fecal-oral route or through respiratory secretions.
As an example, many Gram-negative bacteria possess R (resistance) plasmids that have genes coding for multiple antibiotic resistance through the mechanisms stated above, as well as transfer genes coding for a sex pilus. Such an organism can conjugate with other bacteria and transfer an R plasmid to them. Escherichia coli, Proteus, Serratia, Salmonella, Shigella, and Pseudomonas are examples of bacteria that frequently have R plasmids. Because of the problem of antibiotic resistance, antibiotic susceptibility testing is usually done in the clinical laboratory to determine which antimicrobial chemotherapeutic agents will most likely be effective on a particular strain of microorganism. This is discussed in the next section.
To illustrate how plasmids carrying genes coding for antibiotic resistance can be picked up by antibiotic-sensitive bacteria, in today’s lab we will use plasmid DNA to transform an Escherichia coli sensitive to the antibiotic ampicillin into one that is resistant to the drug.
The E. coli. will be rendered more “competent” to take up plasmid DNA (pAMP), which contains a gene coding for ampicillin resistance, by treating them with a solution of calcium chloride, cold incubation, and a brief heat shock. They will then be plated on 2 types of media: Lauria-Bertani agar (LB) and Lauria-Bertani agar with ampicillin (LB/amp). Only E. coli. that have picked up a plasmid coding for ampicillin resistance will be able to form colonies on the LB/amp agar.
Antibiotic Susceptibility Testing
For some microorganisms, susceptibility to chemotherapeutic agents is predictable. However, for many microorganisms (Pseudomonas, Staphylococcus aureus, and Gram-negative enteric bacilli such as Escherichia coli, Serratia, Proteus, etc.) there is no reliable way of predicting which antimicrobial agent will be effective in a given case. This is especially true with the emergence of many antibiotic-resistant strains of bacteria. Because of this, antibiotic susceptibility testing is often essential in order to determine which antimicrobial agent to use against a specific strain of bacterium.
Several tests may be used to tell a physician which antimicrobial agent is most likely to combat a specific pathogen:
Tube dilution tests
In this test, a series of culture tubes are prepared, each containing a liquid medium and a different concentration of a chemotherapeutic agent. The tubes are then inoculated with the test organism and incubated for 16–20 hours at 35°C. After incubation, the tubes are examined for turbidity (growth). The lowest concentration of chemotherapeutic agent capable of preventing growth of the test organism is the minimum inhibitory concentration (MIC).
Subculturing of tubes showing no turbidity into tubes containing medium, but no chemotherapeutic agent, can determine the minimum bactericidal concentration (MBC). MBC is the lowest concentration of the chemotherapeutic agent that results in no growth (turbidity) of the subcultures. These tests, however, are rather time-consuming and expensive to perform.
The agar diffusion test (Bauer-Kirby test)
A procedure commonly used in clinical labs to determine antimicrobial susceptibility is the Bauer-Kirby disc diffusion method. In this test, the in vitro response of bacteria to a standardized antibiotic-containing disc has been correlated with the clinical response of patients given that drug.
In the development of this method, a single high-potency disc of each chosen chemotherapeutic agent was used. Zones of growth inhibition surrounding each type of disc were correlated with the minimum inhibitory concentrations of each antimicrobial agent (as determined by the tube dilution test). The MIC for each agent was then compared to the usually attained blood level in the patient with adequate dosage. Categories of “Resistant,” “Intermediate,” and “Sensitive” were then established.
The basic steps for the Bauer-Kirby method of antimicrobial susceptibility testing are:
- Prepare a standard turbidity inoculum of the test bacterium so that a certain density of bacteria will be put on the plate.
- Inoculate a 150-mm Mueller-Hinton agar plate with the standardized inoculum, so as to cover the entire agar surface with bacteria.
- Place standardized antibiotic-containing discs on the plate.
- Incubate the plate at 35°C for 18–20 hours.
- Measure the diameter of any resulting zones of inhibition in millimeters (mm).
- Determine if the bacterium is susceptible, moderately susceptible, intermediate, or resistant to each antimicrobial agent.
The term intermediate generally means that the result is inconclusive for that drug-organism combination. The term moderately susceptible is usually applied to those situations where a drug may be used for infections in a particular body site, e.g., cystitis, because the drug becomes highly concentrated in the urine.
Automated tests
Computerized automated tests have been developed for antimicrobial susceptibility testing. These tests measure the inhibitory effect of the antimicrobial agents in a liquid medium by using light-scattering to determine growth of the test organism. Results can be obtained within a few hours. Labs performing very large numbers of susceptibility tests frequently use the automated methods, but the equipment is quite expensive.
Procedures
Microbial Resistance to Antimicrobial Chemotherapeutic Agents
Materials
Plasmid DNA (pAMP) on ice, calcium chloride solution on ice, 2 sterile culture tubes, 1 tube of LB broth, 2 plates of LB agar, 2 plates of LB agar with ampicillin (LB/amp), sterile 1-mL transfer pipettes, sterile plastic inoculating loops, bent glass rod, turntable, alcohol, beaker of ice, water bath at 42°C.
Organism
LB agar culture of Escherichia coli
Microbial Resistance Procedure
- Label one LB agar plate “Transformed bacteria, positive control” and the other LB agar plate “Wild-type bacteria, positive control.” Label one LB/amp agar plate “Transformed bacteria, experiment” and the other LB/amp agar plate “Wild-type bacteria, negative control.”
- Label one sterile culture tube “(+)AMP” and the other “(–)AMP.” Using a sterile 1-mL transfer pipette, add 250 µL of ice cold calcium chloride to each tube. Place both tubes on ice. Using a sterile plastic inoculating loop, transfer 1–2 large colonies of E. coli. into the (+)AMP tube and vigorously tap against the wall of the tube to dislodge all the bacteria. Immediately suspend the cells by repeatedly pipetting in and out with a sterile transfer pipette until no visible clumps of bacteria remain. Return the tube to the ice.
- Repeat step 3, this time using the (–)AMP tube and return to the ice.
- Using a sterile plastic inoculating loop, add 1 loopful of pDNA (plasmid DNA) solution to the (+)AMP tube and swish the loop to mix the DNA. Return to the ice.
- Incubate both tubes on ice for 15 minutes.
- After 15 minutes, “heat-shock” both tube of bacteria by immersing them in a 42°C water bath for 90 seconds. Return both tubes to the ice for 1 minute or more.
- Using a sterile 1-mL transfer pipette, add 250 µL of LB broth to each tube. Tap tubes with your fingers to mix. Set tubes in a test tube rack at room temperature.
- Using a sterile 1-mL transfer pipette, add 100 mL of E. coli. suspension from the (–)AMP tube onto the LB/amp agar plate labeled “Wild-type bacteria, negative control.” Add another 100 L of E. coli. from the (–)AMP to the LB agar plate labeled “Wild-type bacteria, positive control.”
- Using a bent glass rod dipped in alcohol and flamed, spread the bacteria thoroughly over both agar plates. Make sure you reflame the glass rod between plates.
- Using a sterile 1-mL transfer pipette, add 100 mL of E. coli. suspension from the (+)AMP tube onto the LB/amp agar plate labeled “Transformed bacteria, experiment.” Add another 100 L of E. coli. from the (+)AMP to the LB agar plate labeled “Transformed bacteria, positive control.”
- Immediately spread as in step 10.
- Incubate all plates at 37°C.
Antibiotic Susceptibility Testing
Materials
- 150-mm Mueller-Hinton agar plates (3)
- Sterile swabs (3)
- An antibiotic disc dispenser containing discs of antibiotics commonly effective against Gram-positive bacteria, and 1 containing discs of antibiotics commonly effective against Gram-negative bacteria.
Organisms
- Trypticase soy broth cultures of Staphylococcus aureus (Gram-positive)
- Escherichia coli (Gram-negative), and Pseudomonas aeruginosa (Gram-negative)
Antibiotic Susceptibility Testing Procedure
- Take 3 Mueller-Hinton agar plates. Label one S. aureus, one E. coli., and one P. aeruginosa.
- Using your wax marker, divide each plate into thirds to guide your streaking.
- Dip a sterile swab into the previously standardized tube of S. aureus. Squeeze the swab against the inner wall of the tube to remove excess liquid.
- Streak the swab perpendicular to each of the 3 lines drawn on the plate, overlapping the streaks to assure complete coverage of the entire agar surface with inoculum.
- Repeat steps 3 and 4 for the E. coli. and P. aeruginosa plates.
- Using the appropriate antibiotic disc dispenser, place Gram-positive antibiotic-containing discs on the plate of S. aureus and Gram-negative antibiotic-containing discs on the plates of E. coli. and P. aeruginosa.
- Make sure that one of each of the antibiotic-containing discs in the dispenser is on the plate, and touch each disc lightly with sterile forceps to make sure it adheres to the agar surface.
- Incubate the 3 plates upside-down at 37°C until the next lab period.
- Using a metric ruler, measure the diameter of the zone of inhibition around each disc on each plate in mm by placing the ruler on the bottom of the plate
- Determine whether each organism is susceptible, moderately susceptible, intermediate, or resistant to each chemotherapeutic agent using the standardized table, and record your results.
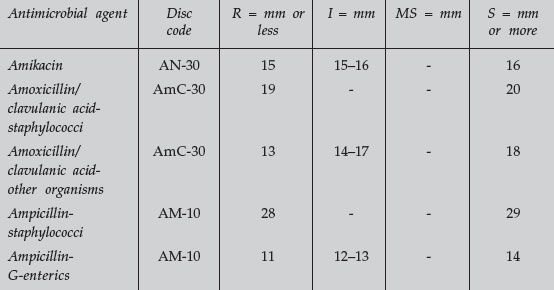 TABLE 1 Zone Size Interpretive Chart for Bauer-Kirby Test
TABLE 1 Zone Size Interpretive Chart for Bauer-Kirby Test
Microbial Resistance to Antimicrobial Chemotherapeutic Agents
Count the number of colonies on each plate. If the growth is too dense to count individual colonies, record “lawn” (bacteria cover nearly the entire agar surface).
Antibiotic Susceptibility Testing: Bauer-Kirby Method
Interpret the results following steps 9 and 10 of the procedure and record your results.
Performance Objectives
Antimicrobial Chemotherapeutic Agents
- Define the following: antibiotic, antimicrobial chemotherapeutic chemical, narrow-spectrum antibiotic, broad-spectrum antibiotic.
- Discuss the meaning of selective toxicity in terms of antimicrobial chemotherapy.
- List 4 genera of microorganisms that produce useful antibiotics.
- Describe 4 different major modes of action of antimicrobial chemotherapeutic chemicals, and name 3 examples of drugs fitting each mode of action.
Microbial Resistance to Antimicrobial Agents
Discussion
- Describe 5 mechanisms by which microorganisms may resist antimicrobial chemotherapeutic agents.
- Briefly describe R plasmids and name 4 bacteria that commonly possess these plasmids.
Results
Interpret the results of the Escherichia coli plasmid transformation experiment.
Antibiotic Susceptibility Testing
Discussion
- Explain why antimicrobial susceptibility testing is often essential in choosing the proper chemotherapeutic agent for use in treating an infection.
- Define MIC.
Results
Interpret the results of a Bauer-Kirby antimicrobial susceptibility test when given a Mueller-Hinton agar plate, a metric ruler, and a standardized zone-size interpretation table.




