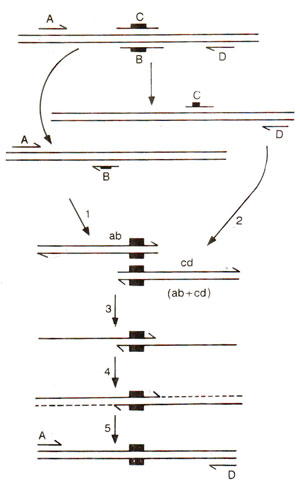
Different schemes of PCR
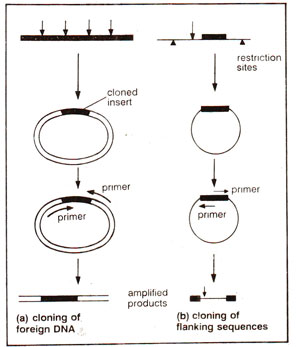
Fig. 39.21. Inverse PCR : (a) use of vector border sequences as primers for amplification of inserted sequence, (b) use of border sequences of a gene to amplify its flanking regulatory sequences. (Modified and redrawn from Dharmalingam, K.-Biospectra, Nov. - Dec, 1990 page 3-8).
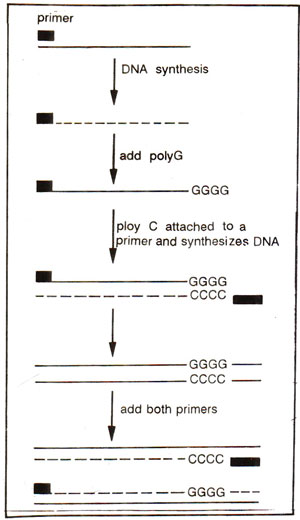
Fig. 39.22. Anchored PCR, using only one primer at the initial stage of amplification. (Modified and redrawn from Dharmalingam, K.-Biospectra, Nov, -Dec, 1990 page 3-8).
Inverse PCR. In this technique, amplification of those DNA sequences takes place, which are away from the primers and not those which are flanked by the primers. For instance if the border sequences of a DNA segment are not known and those of a vector are known, then the sequence to be amplified may be cloned in the vector and border sequences of vector may be used as primers in such a way that the polymerization proceed in inverse direction i.e. towards the inserted segment, and not away from it towards the DNA sequence of vector from which primers have been derived (Fig. 39.21a). Similarly, if the gene sequence is known, one can use its border sequences as primers to amplify the sequences flanking this gene e.g. the regulatory sequences (Fig. 39.21b).
Anchored PCR. In the basic PCR technique and the inverse PCR, one has to use two primers representing the sequences lying at both ends of sequence to be amplified. But sometimes, we may have knowledge about the sequence at only one of the two ends of the DNA sequence to be amplified. In such cases anchored PCR may be used, which will utilize only one primer instead of two primers. In this technique, due to the use of one primer, only one strand will be copied first, after which a poly G will be attached at the end of the newly synthesized strand. This newly synthesized strand with poly G tail at its 3' end will then become template for the daughter strand synthesis utilizing an anchor primer with which a poly C sequence is linked to complement with poly G of the template. In the next cycle, both the original primer and anchored primer will be used for gene amplification (Fig. 39.22).

Fig. 39.21. Inverse PCR : (a) use of vector border sequences as primers for amplification of inserted sequence, (b) use of border sequences of a gene to amplify its flanking regulatory sequences. (Modified and redrawn from Dharmalingam, K.-Biospectra, Nov. - Dec, 1990 page 3-8).

Fig. 39.22. Anchored PCR, using only one primer at the initial stage of amplification. (Modified and redrawn from Dharmalingam, K.-Biospectra, Nov, -Dec, 1990 page 3-8).
Extension of these primers results in the formation of a complex gene, with the mutation incorporated into it (Fig. 39.24).