Tetrahydrobenzylisoquinoline Alkaloid Biosynthesis
On the pathway leading from L-tyrosine to the first tetrahydrobenzylisoquinoline
alkaloidal intermediate (S)-norcoclaurine, cDNAs encoding tyrosine/dopa decarboxylases
(
tydc) have been isolated (Fig. 10.5) (Facchini and De Luca, 1994).
The transformation of (S)-norcoclaurine to the central isoquinoline alkaloid biosynthetic
intermediate (S)-reticuline is quite well understood at both the enzyme and gene level (Fig. 10.6). (S)-Norcoclaurine is O-methylated at the 6-position by
(R,S)-norcoclaurine 6-O-methyltransferase (6-
omt) (Ru¨ ffer
et al., 1983). cDNAs
encoding this enzyme have been isolated from
Thalictrum tuberosum,
Coptis japonica,
and
P. somniferum (Frick and Kutchan, 1999; Morishige
et al., 2000; Ounaroon
et al., 2003). (S)-
Coclaurine is next N-methylated by (R,S)-coclaurine
N-methyltransferase (Frenzel and Zenk, 1990a). This cDNA has been characterized
from
C. japonica (Choi
et al., 2002) and
P. somniferum (S. Haase, J. Ziegler,
S. Frick, and T. M. Kutchan, unpublished
data). (S)-N-Methylcoclaurine is
hydroxylated by the cytochrome
P450 dependent monooxygenase
cyp80b1
(S)-N-methylcoclaurine 3'-hydroxlyase (Pauli and Kutchan, 1998). The cDNA
encoding this cytochrome P450 has been isolated from the California poppy
Eschscholzia californica and from
P. somniferum (Huang and Kutchan, 2000;
Pauli and Kutchan, 1998). (S)-3'-Hydroxy-N-methylcoclaurine is methylated
to (S)-reticuline by (R,S)-3'-hydroxy-N-methylcoclaurine 4'-O-methyltransferase
(4'-
omt) (Frenzel and Zenk, 1990b). The cDNA 4'-
omt has been isolated from
C. japonica (Morishige
et al., 2000) and from
P. somniferum (Ziegler
et al., 2005).
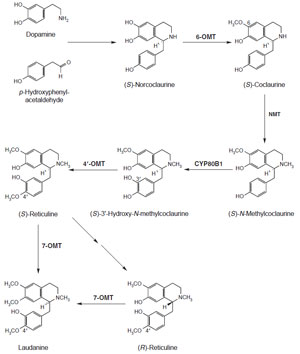 |
| FIGURE 10.6 Schematic representation of the
biosynthetic pathway leading from dopamine and
p-hydroxyphenylacetaldehyde to laudanine.
6-omt, (R,S)-norcoclaurine
6-O-methyltransferase;
NMT, (R,S)-coclaurine,
N-methyltransferase; cyp80b1,
(S)-N-methylcoclaurine 3'-hydroxylase;
4'-omt, (R,S)-3'-hydroxy-N-methylcoclaurine
4'-O-methyltransferase; 7-omt,
(R,S)-reticuline
7-O-methyltransferase. |
(S)-Reticuline is the chemical chameleon of isoquinoline alkaloid biosynthesis,
which can lead to a plethora of alkaloidal structures. In
P. somniferum,
(R,S)-reticuline can be methylated by (R,S)-reticuline 7-O-methyltransferase, for
which the cDNA 7-
omt has been described, to the tetrahydrobenzylisoquinoline
laudanine (Fig. 10.6 Ounaroon
et al., 2003). Along the pathway in which
(S)-reticuline is specifically converted to morphine, cDNAs encoding two biosynthetic
enzymes have been identified (Fig. 10.7). Salutaridinol 7-O-acetyltransferase,
encoded by
SalAT, transfers an acetyl moiety from acetyl-CoA to the 7-hydroxyl
group of salutaridinol (Grothe
et al., 2001; Lenz and Zenk, 1995a). Codeinone
reductase is encoded by
cor1 and catalyzes the penultimate step in morphine
biosynthesis, the NADPH-dependent reduction of the keto moiety of
codeinone
to the 6-hydroxyl group of codeine (Lenz and Zenk, 1995b; Unterlinner
et
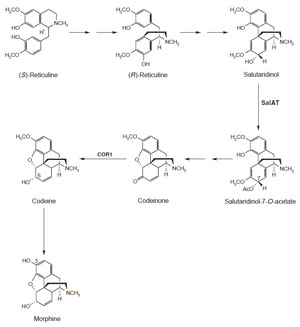 |
| FIGURE 10.7 Schematic representation of the
biosynthetic pathway leading from (S)-reticuline
to morphine. SalAT, salutaridinol
7-O-acetyltransferase; COR1, codeinone
reductase. |
al., 1999).
In
P. somniferum and
E. californica,
the N-methyl group of (S)-reticuline can be
oxi
datively cyclized by the
bbe to the bridge carbon, C-8, of (S)-scoulerine (Fig. 10.8; Rink and Böhm, 1975; Steffens
et al., 1985). (S)-Scoulerine is then further
converted in these plants to antimicrobial benzo[c]phenathridine alkaloids, such
as sanguinarine. cDNAs encoding the
bbe have been isolated from
E. californica,
P. somniferum, and
Berberis stolonifera (Chou and Kutchan, 1998; Dittrich and
Kutchan, 1991; Facchini
et al., 1996; Huang and Kutchan, 2000). (S)-Reticuline
is converted via (S)-scoulerine to berberine alkaloids in
Berberis and
Coptis species. Along the biosynthetic pathway to berberine, two cDNAs have
been identified from
C. japonica. (S)-Scoulerine is methylated by (S)-scoulerine
9-O-methyltransferase (9-
omt) (Muemmler
et al., 1985;
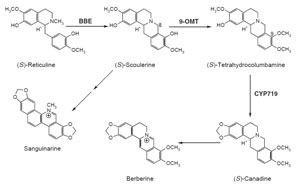 |
| FIGURE 10.8 Schematic representation of the
biosynthetic pathway leading from (S)-reticuline
to sanguinarine and berberine. bbe, berberine
bridge enzyme; 9-omt, (S)-scoulerine
9-O-methyltransferase; CYP719,
(S)-canadine synthase. |
Takeshita
et al., 1995) to
(S)-tetrahydrocolumbamine which is subsequently acted upon by CYP719 (Bauer
and Zenk, 1991; Ikezawa
et al., 2003; Rueffer and Zenk, 1994), a cytochrome
P450-
dependent enzyme that catalyzes formation of the methylenedioxy bridge of
(S)-canadine.
With the current collection of cDNAs encoding enzymes of tetrahydrobenzylisoquinoline
alkaloid biosynthesis, some progress has also been made with
respect to our understanding of the spatial regulation of this biosynthesis.
The cellular localization of tetrahydrobenzylisoquinoline alkaloid biosynthesis
will next be considered.







